Integrating Full-Length Transcriptome and RNA Sequencing of Siberian Wildrye (Elymus sibiricus) to Reveal Molecular Mechanisms in Response to Drought Stress
- PMID: 37514333
- PMCID: PMC10385362
- DOI: 10.3390/plants12142719
Integrating Full-Length Transcriptome and RNA Sequencing of Siberian Wildrye (Elymus sibiricus) to Reveal Molecular Mechanisms in Response to Drought Stress
Abstract
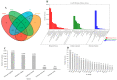
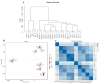
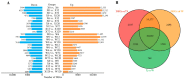
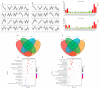
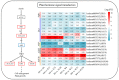
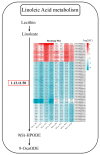
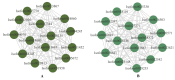
Drought is one of the most significant limiting factors affecting plant growth and development on the Qinghai-Tibet Plateau (QTP). Mining the drought-tolerant genes of the endemic perennial grass of the QTP, Siberian wildrye (Elymus sibiricus), is of great significance to creating new drought-resistant varieties which can be used in the development of grassland livestock and restoring natural grassland projects in the QTP. To investigate the transcriptomic responsiveness of E. sibiricus to drought stress, PEG-induced short- and long-term drought stress was applied to two Siberian wildrye genotypes (drought-tolerant and drought-sensitive accessions), followed by third- and second-generation transcriptome sequencing analysis. A total of 40,708 isoforms were detected, of which 10,659 differentially expressed genes (DEGs) were common to both genotypes. There were 2107 and 2498 unique DEGs in the drought-tolerant and drought-sensitive genotypes, respectively. Additionally, 2798 and 1850 DEGs were identified in the drought-tolerant genotype only under short- and long-term conditions, respectively. DEGs numbering 1641 and 1330 were identified in the drought-sensitive genotype only under short- and long-term conditions, respectively. Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis revealed that all the DEGs responding to drought stress in E. sibiricus were mainly associated with the mitogen-activated protein kinase (MAKP) signaling pathway, plant hormone signal transduction, the linoleic acid metabolism pathway, the ribosome pathway, and plant circadian rhythms. In addition, Nitrate transporter 1/Peptide transporter family protein 3.1 (NPF3.1) and Auxin/Indole-3-Acetic Acid (Aux/IAA) family protein 31(IAA31) also played an important role in helping E. sibiricus resist drought. This study used transcriptomics to investigate how E. sibiricus responds to drought stress, and may provide genetic resources and references for research into the molecular mechanisms of drought resistance in native perennial grasses and for breeding drought-tolerant varieties.
Keywords: Elymus sibiricus; WGCNA; comparative transcriptomics; drought stress; endemic forage.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Comparative Metabolomic Studies of Siberian Wildrye (Elymus sibiricus L.): A New Look at the Mechanism of Plant Drought Resistance.Int J Mol Sci. 2022 Dec 27;24(1):452. doi: 10.3390/ijms24010452. Int J Mol Sci. 2022. PMID: 36613896 Free PMC article.
-
Comparative physiological and transcriptomic analyses reveal genotype specific response to drought stress in Siberian wildrye (Elymus sibiricus).Sci Rep. 2024 Sep 10;14(1):21060. doi: 10.1038/s41598-024-71847-9. Sci Rep. 2024. PMID: 39256456 Free PMC article.
-
Transcriptome profiles identify the common responsive genes to drought stress in two Elymus species.J Plant Physiol. 2020 Jul;250:153183. doi: 10.1016/j.jplph.2020.153183. Epub 2020 May 11. J Plant Physiol. 2020. PMID: 32422512
-
Molecular Phylogeography and Intraspecific Divergences in Siberian Wildrye (Elymus sibiricus L.) Wild Populations in China, Inferred From Chloroplast DNA Sequence and cpSSR Markers.Front Plant Sci. 2022 May 19;13:862759. doi: 10.3389/fpls.2022.862759. eCollection 2022. Front Plant Sci. 2022. PMID: 35665183 Free PMC article.
-
Metabolomic Analysis of Elymus sibiricus Exposed to UV-B Radiation Stress.Molecules. 2024 Oct 30;29(21):5133. doi: 10.3390/molecules29215133. Molecules. 2024. PMID: 39519780 Free PMC article.
Cited by
-
Extensive transcriptome data providing great efficacy in genetic research and adaptive gene discovery: a case study of Elymus sibiricus L. (Poaceae, Triticeae).Front Plant Sci. 2024 Sep 19;15:1457980. doi: 10.3389/fpls.2024.1457980. eCollection 2024. Front Plant Sci. 2024. PMID: 39363927 Free PMC article.
References
-
- Zhao Z.L., Zhang Y.L., Liu F.G., Zhang H.F., Zhou Q., Liu P., Zou X.H. Drought disaster risk analysis of Tibetan Plateau. J. Mt. Sci. 2013;31:672–684. doi: 10.3969/j.issn.1008-2786.2013.06.005. - DOI
Grants and funding
- 2022YFQ0076/Sichuan Provincial Regional Innovation Cooperation Project
- 31772661/National Natural Science Foundation of China
- 20220102/Emergency Sci. & Tech Project from National Forestry and Grassland Administration ('Breeding and Cultivation of Fine Herbage Varieties')
- CARS-34/National Modern Forages Industry Technology System
- 2021YFYZ0013-2/Sichuan Provincial Key Research Project for Forages Breeding
LinkOut - more resources
Full Text Sources

