Investigating the Prevalence of RNA-Binding Metabolic Enzymes in E. coli
- PMID: 37511294
- PMCID: PMC10380284
- DOI: 10.3390/ijms241411536
Investigating the Prevalence of RNA-Binding Metabolic Enzymes in E. coli
Abstract
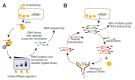
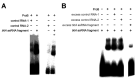
An open research field in cellular regulation is the assumed crosstalk between RNAs, metabolic enzymes, and metabolites, also known as the REM hypothesis. High-throughput assays have produced extensive interactome data with metabolic enzymes frequently found as hits, but only a few examples have been biochemically validated, with deficits especially in prokaryotes. Therefore, we rationally selected nineteen Escherichia coli enzymes from such datasets and examined their ability to bind RNAs using two complementary methods, iCLIP and SELEX. Found interactions were validated by EMSA and other methods. For most of the candidates, we observed no RNA binding (12/19) or a rather unspecific binding (5/19). Two of the candidates, namely glutamate-5-kinase (ProB) and quinone oxidoreductase (QorA), displayed specific and previously unknown binding to distinct RNAs. We concentrated on the interaction of QorA to the mRNA of yffO, a grounded prophage gene, which could be validated by EMSA and MST. Because the physiological function of both partners is not known, the biological relevance of this interaction remains elusive. Furthermore, we found novel RNA targets for the MS2 phage coat protein that served us as control. Our results indicate that RNA binding of metabolic enzymes in procaryotes is less frequent than suggested by the results of high-throughput studies, but does occur.
Keywords: MS2 phage; REM hypothesis; RNA-binding protein; SELEX; metabolic enzymes; prokaryotes; quinone oxidoreductase; unconventional RNA binding.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

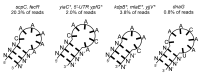


Similar articles
-
Oxidative-stress-inducible qorA encodes an NADPH-dependent quinone oxidoreductase catalysing a one-electron reduction in Staphylococcus aureus.Microbiology (Reading). 2003 Feb;149(Pt 2):389-398. doi: 10.1099/mic.0.25796-0. Microbiology (Reading). 2003. PMID: 12624201
-
Analysis of phage MS2 coat protein mutants expressed from a reconstituted phagemid reveals that proline 78 is essential for viral infectivity.J Mol Biol. 1997 Feb 14;266(1):1-7. doi: 10.1006/jmbi.1996.0786. J Mol Biol. 1997. PMID: 9054964
-
Interactions of Escherichia coli RNA with bacteriophage MS2 coat protein: genomic SELEX.Nucleic Acids Res. 2000 Nov 1;28(21):E93. doi: 10.1093/nar/28.21.e93. Nucleic Acids Res. 2000. PMID: 11058143 Free PMC article.
-
[Interaction of RNA-containing bacteriophages with the host cells: MS2-induced E. coli mutants and formation of DNA-containing derivatives of MS2 bacteriophage].Tsitol Genet. 2008 Jan-Feb;42(1):73-90. Tsitol Genet. 2008. PMID: 18411761 Review. Russian.
-
The mutual interaction of glycolytic enzymes and RNA in post-transcriptional regulation.RNA. 2022 Nov;28(11):1446-1468. doi: 10.1261/rna.079210.122. Epub 2022 Aug 16. RNA. 2022. PMID: 35973722 Free PMC article. Review.
Cited by
-
Iron-sulfur protein odyssey: exploring their cluster functional versatility and challenging identification.Metallomics. 2024 May 2;16(5):mfae025. doi: 10.1093/mtomcs/mfae025. Metallomics. 2024. PMID: 38744662 Free PMC article. Review.
References
-
- Hacisuleyman E., Goff L.A., Trapnell C., Williams A., Henao-Mejia J., Sun L., McClanahan P., Hendrickson D.G., Sauvageau M., Kelley D.R., et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat. Struct. Mol. Biol. 2014;21:198–206. doi: 10.1038/nsmb.2764. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

