The HDAC6-RNF168 axis regulates H2A/H2A.X ubiquitination to enable double-strand break repair
- PMID: 37503842
- PMCID: PMC10516627
- DOI: 10.1093/nar/gkad631
The HDAC6-RNF168 axis regulates H2A/H2A.X ubiquitination to enable double-strand break repair
Abstract
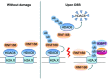
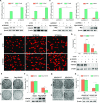
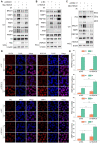
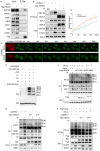
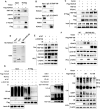
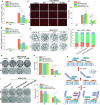
Histone deacetylase 6 (HDAC6) mediates DNA damage signaling by regulating the mismatch repair and nucleotide excision repair pathways. Whether HDAC6 also mediates DNA double-strand break (DSB) repair is unclear. Here, we report that HDAC6 negatively regulates DSB repair in an enzyme activity-independent manner. In unstressed cells, HDAC6 interacts with H2A/H2A.X to prevent its interaction with the E3 ligase RNF168. Upon sensing DSBs, RNF168 rapidly ubiquitinates HDAC6 at lysine 116, leading to HDAC6 proteasomal degradation and a restored interaction between RNF168 and H2A/H2A.X. H2A/H2A.X is ubiquitinated by RNF168, precipitating the recruitment of DSB repair factors (including 53BP1 and BRCA1) to chromatin and subsequent DNA repair. These findings reveal novel regulatory machinery based on an HDAC6-RNF168 axis that regulates the H2A/H2A.X ubiquitination status. Interfering with this axis might be leveraged to disrupt a key mechanism of cancer cell resistance to genotoxic damage and form a potential therapeutic strategy for cancer.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


Similar articles
-
Ubiquitin-induced RNF168 condensation promotes DNA double-strand break repair.Proc Natl Acad Sci U S A. 2024 Jul 9;121(28):e2322972121. doi: 10.1073/pnas.2322972121. Epub 2024 Jul 5. Proc Natl Acad Sci U S A. 2024. PMID: 38968116 Free PMC article.
-
USP14 regulates DNA damage repair by targeting RNF168-dependent ubiquitination.Autophagy. 2018;14(11):1976-1990. doi: 10.1080/15548627.2018.1496877. Epub 2018 Aug 10. Autophagy. 2018. PMID: 29995557 Free PMC article.
-
New answers to the old RIDDLE: RNF168 and the DNA damage response pathway.FEBS J. 2022 May;289(9):2467-2480. doi: 10.1111/febs.15857. Epub 2021 Apr 16. FEBS J. 2022. PMID: 33797206 Free PMC article. Review.
-
RNF168-mediated H2A neddylation antagonizes ubiquitylation of H2A and regulates DNA damage repair.J Cell Sci. 2014 May 15;127(Pt 10):2238-48. doi: 10.1242/jcs.138891. Epub 2014 Mar 14. J Cell Sci. 2014. PMID: 24634510
-
Opposing roles of RNF8/RNF168 and deubiquitinating enzymes in ubiquitination-dependent DNA double-strand break response signaling and DNA-repair pathway choice.J Radiat Res. 2016 Aug;57 Suppl 1(Suppl 1):i33-i40. doi: 10.1093/jrr/rrw027. Epub 2016 Mar 16. J Radiat Res. 2016. PMID: 26983989 Free PMC article. Review.
Cited by
-
Dual inhibitors of DNMT and HDAC induce viral mimicry to induce antitumour immunity in breast cancer.Cell Death Discov. 2024 Mar 15;10(1):143. doi: 10.1038/s41420-024-01895-7. Cell Death Discov. 2024. PMID: 38490978 Free PMC article.
-
RNF126, 168 and CUL1: The Potential Utilization of Multi-Functional E3 Ubiquitin Ligases in Genome Maintenance for Cancer Therapy.Biomedicines. 2023 Sep 13;11(9):2527. doi: 10.3390/biomedicines11092527. Biomedicines. 2023. PMID: 37760968 Free PMC article. Review.
-
Chromatin lysine acylation: On the path to chromatin homeostasis and genome integrity.Cancer Sci. 2024 Nov;115(11):3506-3519. doi: 10.1111/cas.16321. Epub 2024 Aug 18. Cancer Sci. 2024. PMID: 39155589 Free PMC article. Review.
-
The role of ubiquitination in health and disease.MedComm (2020). 2024 Sep 25;5(10):e736. doi: 10.1002/mco2.736. eCollection 2024 Oct. MedComm (2020). 2024. PMID: 39329019 Free PMC article. Review.
-
The Function of H2A Histone Variants and Their Roles in Diseases.Biomolecules. 2024 Aug 12;14(8):993. doi: 10.3390/biom14080993. Biomolecules. 2024. PMID: 39199381 Free PMC article. Review.
References
-
- Lord C.J., Ashworth A.. The DNA damage response and cancer therapy. Nature. 2012; 481:287–294. - PubMed
-
- Panier S., Durocher D. Push back to respond better: regulatory inhibition of the DNA double-strand break response. Nat. Rev. Mol. Cell Biol. 2013; 14:661–672. - PubMed
-
- Zhang K., Ning Y., Kong F., Chen X., Cai Y.. Genome instability in pathogenesis of tuberculosis. Genome Instab. Dis. 2021; 2:331–338.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

