This is a preprint.
ACKR3-arrestin2/3 complexes reveal molecular consequences of GRK-dependent barcoding
- PMID: 37502840
- PMCID: PMC10370059
- DOI: 10.1101/2023.07.18.549504
ACKR3-arrestin2/3 complexes reveal molecular consequences of GRK-dependent barcoding
Abstract
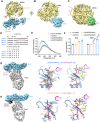
Atypical chemokine receptor 3 (ACKR3, also known as CXCR7) is a scavenger receptor that regulates extracellular levels of the chemokine CXCL12 to maintain responsiveness of its partner, the G protein-coupled receptor (GPCR), CXCR4. ACKR3 is notable because it does not couple to G proteins and instead is completely biased towards arrestins. Our previous studies revealed that GRK2 and GRK5 install distinct distributions of phosphates (or "barcodes") on the ACKR3 carboxy terminal tail, but how these unique barcodes drive different cellular outcomes is not understood. It is also not known if arrestin2 (Arr2) and 3 (Arr3) bind to these barcodes in distinct ways. Here we report cryo-electron microscopy structures of Arr2 and Arr3 in complex with ACKR3 phosphorylated by either GRK2 or GRK5. Unexpectedly, the finger loops of Arr2 and 3 directly insert into the detergent/membrane instead of the transmembrane core of ACKR3, in contrast to previously reported "core" GPCR-arrestin complexes. The distance between the phosphorylation barcode and the receptor transmembrane core regulates the interaction mode of arrestin, alternating between a tighter complex for GRK5 sites and heterogenous primarily "tail only" complexes for GRK2 sites. Arr2 and 3 bind at different angles relative to the core of ACKR3, likely due to differences in membrane/micelle anchoring at their C-edge loops. Our structural investigations were facilitated by Fab7, a novel Fab that binds both Arr2 and 3 in their activated states irrespective of receptor or phosphorylation status, rendering it a potentially useful tool to aid structure determination of any native GPCR-arrestin complex. The structures provide unprecedented insight into how different phosphorylation barcodes and arrestin isoforms can globally affect the configuration of receptor-arrestin complexes. These differences may promote unique downstream intracellular interactions and cellular responses. Our structures also suggest that the 100% bias of ACKR3 for arrestins is driven by the ability of arrestins, but not G proteins, to bind GRK-phosphorylated ACKR3 even when excluded from the receptor cytoplasmic binding pocket.
Figures





Similar articles
-
Atypical Chemokine Receptor 3 "Senses" CXC Chemokine Receptor 4 Activation Through GPCR Kinase Phosphorylation.Mol Pharmacol. 2023 Oct;104(4):174-186. doi: 10.1124/molpharm.123.000710. Epub 2023 Jul 20. Mol Pharmacol. 2023. PMID: 37474305 Free PMC article.
-
Atypical Chemokine Receptor 3 'Senses' CXC Chemokine Receptor 4 Activation Through GPCR Kinase Phosphorylation.bioRxiv [Preprint]. 2023 Mar 10:2023.02.25.530029. doi: 10.1101/2023.02.25.530029. bioRxiv. 2023. Update in: Mol Pharmacol. 2023 Oct;104(4):174-186. doi: 10.1124/molpharm.123.000710. PMID: 36865154 Free PMC article. Updated. Preprint.
-
Differential Involvement of ACKR3 C-Tail in β-Arrestin Recruitment, Trafficking and Internalization.Cells. 2021 Mar 11;10(3):618. doi: 10.3390/cells10030618. Cells. 2021. PMID: 33799570 Free PMC article.
-
G protein-coupled receptor interactions with arrestins and GPCR kinases: The unresolved issue of signal bias.J Biol Chem. 2022 Sep;298(9):102279. doi: 10.1016/j.jbc.2022.102279. Epub 2022 Jul 19. J Biol Chem. 2022. PMID: 35863432 Free PMC article. Review.
-
Emerging Roles of the Atypical Chemokine Receptor 3 (ACKR3) in Cardiovascular Diseases.Front Endocrinol (Lausanne). 2022 Jun 29;13:906586. doi: 10.3389/fendo.2022.906586. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 35846294 Free PMC article. Review.
References
-
- Balkwill F. (2004). The significance of cancer cell expression of the chemokine receptor CXCR4. Semin Cancer Biol 14, 171–179. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
