In-silico mining to glean SNPs of pharmaco-clinical importance: an investigation with reference to the Indian populated SNPs
- PMID: 37484779
- PMCID: PMC10356698
- DOI: 10.1007/s40203-023-00154-4
In-silico mining to glean SNPs of pharmaco-clinical importance: an investigation with reference to the Indian populated SNPs
Abstract
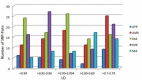
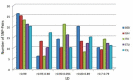
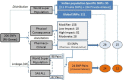
Drugs pharmacology is defined by pharmacokinetics and pharmacodynamics and both of them are affected by genetic variability. Genetic variability varies from population to population, and sometimes even within the population, it exists. Single nucleotide polymorphisms (SNPs) are one of the major genetic variability factors which are found to be associated with the pharmacokinetics and pharmacodynamics process of a drug and are responsible for variable drug response and clinical phenotypes. Studies of SNPs can help to perform genome-wide association studies for their association with pharmacological and clinical events, at the same time; their information can direct genome-wide association studies for their use as biomarkers. With the aim to mine and characterize Indian populated SNPs of pharmacological and clinical importance. Two hundred six candidate SNPs belonging to 43 genes were retrieved from Indian Genome Variation Database. The distribution pattern of considered SNPs was observed against all five world super-populations (AFR, AMR, EAS, EUR, and SAS). Further, their annotation was done through SNP-nexus by considering Human genome reference builds - hg38, pharmacological and clinical information was supplemented by PharmGKB and ClinVar database. At last, to find out the association between SNPs linkage disequilibrium was observed in terms of r2. Overall, the study reported 53 pharmaco-clinical active SNPs and found 24 SNP-pairs as potential markers, and recommended their clinical and experimental validation.
Supplementary information: The online version contains supplementary material available at 10.1007/s40203-023-00154-4.
Keywords: Drug response; Haplotype; Linkage disequilibrium; Molecular marker; Single nucleotide polymorphisms.
© The Author(s), under exclusive licence to Springer-Verlag GmbH Germany, part of Springer Nature 2023. Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Conflict of interest statement
Competing interestsThe authors declare no competing interests.
Figures

Similar articles
-
An ancestry informative marker panel design for individual ancestry estimation of Hispanic population using whole exome sequencing data.BMC Genomics. 2019 Dec 30;20(Suppl 12):1007. doi: 10.1186/s12864-019-6333-6. BMC Genomics. 2019. PMID: 31888480 Free PMC article.
-
Phenotype variations affect genetic association studies of degenerative disc disease: conclusions of analysis of genetic association of 58 single nucleotide polymorphisms with highly specific phenotypes for disc degeneration in 332 subjects.Spine J. 2013 Oct;13(10):1309-20. doi: 10.1016/j.spinee.2013.05.019. Epub 2013 Jun 21. Spine J. 2013. PMID: 23792102
-
Evaluating significance of European-associated index SNPs in the East Asian population for 31 complex phenotypes.BMC Genomics. 2023 Jun 13;24(1):324. doi: 10.1186/s12864-023-09425-y. BMC Genomics. 2023. PMID: 37312035 Free PMC article.
-
High-throughput SNP analysis for genetic association studies.Curr Opin Drug Discov Devel. 2003 May;6(3):317-21. Curr Opin Drug Discov Devel. 2003. PMID: 12833663 Review.
-
Pattern-recognition techniques with haplotype analysis in pharmacogenomics.Pharmacogenomics. 2007 Jan;8(1):75-83. doi: 10.2217/14622416.8.1.75. Pharmacogenomics. 2007. PMID: 17187511 Review.
References
LinkOut - more resources
Full Text Sources

