TMT-Based Proteomics Analysis of the Intervention Effect of Orlistat on Polycystic Ovary Syndrome Rats Induced by Letrozole Combined with a High-Fat Diet
- PMID: 37483206
- PMCID: PMC10357523
- DOI: 10.1021/acsomega.3c00578
TMT-Based Proteomics Analysis of the Intervention Effect of Orlistat on Polycystic Ovary Syndrome Rats Induced by Letrozole Combined with a High-Fat Diet
Abstract
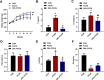
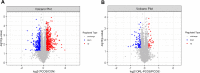
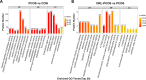
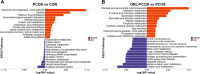
Polycystic ovary syndrome (PCOS) is a complex gynecological endocrine and metabolic disease. Orlistat as a lipase inhibitor may improve the pathological characteristics of PCOS and is the sole antiobesity agent available in various countries. In this study, the PCOS rat models were established using letrozole and high-fat diet. Tandem Mass Tag labeling peptide coupled with liquid chromatography with tandem mass spectrometry (LC-MS/MS) approach was employed to investigate the differentially expressed ovarian proteins (DEPs) in the PCOS and control rats for the effect of PCOS, and in the PCOS and orlistat-treated PCOS rats for the effect of orlistat in PCOS. The orlistat attenuated the body weight gain; decreased the levels of testosterone, luteinizing hormone, a ratio of luteinizing/follicle-stimulating hormones; increased the level of estradiol; and recovered the estrous cycle in PCOS rats. In addition, 795 and 119 DEPs were found in PCOS and orlistat-treated PCOS groups, respectively. Based on the Gene Ontology and Kyoto Encyclopedia of Gene and Genomes pathway analysis of DEPs, orlistat restored the disturbed metabolism of linoleic acid, arachidonic acid, galactose, and glycerolipids, and then improved the chronic inflammation in PCOS rats. This study analyzed the ovarian proteome of orlistat-treated PCOS rats and identified targeted proteins, which explored the pathogenesis of PCOS and the potential effects of orlistat in PCOS rats.
© 2023 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


Similar articles
-
Integrated fecal microbiota and metabolomics analysis of the orlistat intervention effect on polycystic ovary syndrome rats induced by letrozole combined with a high-fat diet.J Ovarian Res. 2023 Jun 5;16(1):109. doi: 10.1186/s13048-023-01193-3. J Ovarian Res. 2023. PMID: 37277785 Free PMC article.
-
The ameliorative effect of CangFu Daotan Decoction on polycystic ovary syndrome of rodent model is associated with m6A methylation and Wnt/β-catenin pathway.Gynecol Endocrinol. 2023 Dec;39(1):2181637. doi: 10.1080/09513590.2023.2181637. Epub 2023 Feb 23. Gynecol Endocrinol. 2023. PMID: 36822223
-
A Rat Model of Polycystic Ovary Syndrome with Insulin Resistance Induced by Letrozole Combined with High Fat Diet.Med Sci Monit. 2020 May 25;26:e922136. doi: 10.12659/MSM.922136. Med Sci Monit. 2020. PMID: 32448863 Free PMC article.
-
Liuwei Dihuang Pills alleviate the polycystic ovary syndrome with improved insulin sensitivity through PI3K/Akt signaling pathway.J Ethnopharmacol. 2020 Mar 25;250:111965. doi: 10.1016/j.jep.2019.111965. Epub 2019 Jun 8. J Ethnopharmacol. 2020. PMID: 31185267
-
The Role of Antiobesity Agents in the Management of Polycystic Ovary Syndrome.Folia Med (Plovdiv). 2018 Dec 1;60(4):512-520. doi: 10.2478/folmed-2018-0036. Folia Med (Plovdiv). 2018. PMID: 31188761 Review.
Cited by
-
Efficacy and safety of orlistat in male patients with overweight/obesity and hyperuricemia: results of a randomized, double-blind, placebo-controlled trial.Lipids Health Dis. 2024 Mar 11;23(1):77. doi: 10.1186/s12944-024-02047-7. Lipids Health Dis. 2024. PMID: 38468241 Free PMC article. Clinical Trial.
References
-
- Teede H. J.; Misso M. L.; Costello M. F.; Dokras A.; Laven J.; Moran L.; Piltonen T.; Norman R. J.; et al. Recommendations from the international evidence-based guideline for the assessment and management of polycystic ovary syndrome. Hum. Reprod. 2018, 33, 1602–1618. 10.1093/humrep/dey256. - DOI - PMC - PubMed
- Wild R. A. Long-term health consequences of PCOS. Hum. Reprod. Update 2002, 8, 231–241. 10.1093/humupd/8.3.231. - DOI - PubMed
LinkOut - more resources
Full Text Sources

