Antiviral HIV-1 SERINC restriction factors disrupt virus membrane asymmetry
- PMID: 37474505
- PMCID: PMC10359404
- DOI: 10.1038/s41467-023-39262-2
Antiviral HIV-1 SERINC restriction factors disrupt virus membrane asymmetry
Abstract
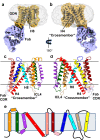
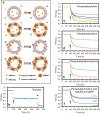
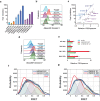
The host proteins SERINC3 and SERINC5 are HIV-1 restriction factors that reduce infectivity when incorporated into the viral envelope. The HIV-1 accessory protein Nef abrogates incorporation of SERINCs via binding to intracellular loop 4 (ICL4). Here, we determine cryoEM maps of full-length human SERINC3 and an ICL4 deletion construct, which reveal that hSERINC3 is comprised of two α-helical bundles connected by a ~ 40-residue, highly tilted, "crossmember" helix. The design resembles non-ATP-dependent lipid transporters. Consistently, purified hSERINCs reconstituted into proteoliposomes induce flipping of phosphatidylserine (PS), phosphatidylethanolamine and phosphatidylcholine. Furthermore, SERINC3, SERINC5 and the scramblase TMEM16F expose PS on the surface of HIV-1 and reduce infectivity, with similar results in MLV. SERINC effects in HIV-1 and MLV are counteracted by Nef and GlycoGag, respectively. Our results demonstrate that SERINCs are membrane transporters that flip lipids, resulting in a loss of membrane asymmetry that is strongly correlated with changes in Env conformation and loss of infectivity.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Beyond Impairment of Virion Infectivity: New Activities of the Anti-HIV Host Cell Factor SERINC5.Viruses. 2024 Feb 12;16(2):284. doi: 10.3390/v16020284. Viruses. 2024. PMID: 38400059 Free PMC article. Review.
-
Potent Enhancement of HIV-1 Replication by Nef in the Absence of SERINC3 and SERINC5.mBio. 2019 Jun 11;10(3):e01071-19. doi: 10.1128/mBio.01071-19. mBio. 2019. PMID: 31186327 Free PMC article.
-
TIM-mediated inhibition of HIV-1 release is antagonized by Nef but potentiated by SERINC proteins.Proc Natl Acad Sci U S A. 2019 Mar 19;116(12):5705-5714. doi: 10.1073/pnas.1819475116. Epub 2019 Mar 6. Proc Natl Acad Sci U S A. 2019. PMID: 30842281 Free PMC article.
-
SERINC5 Potently Restricts Retrovirus Infection In Vivo.mBio. 2020 Jul 14;11(4):e00588-20. doi: 10.1128/mBio.00588-20. mBio. 2020. PMID: 32665269 Free PMC article.
-
SERINC as a Restriction Factor to Inhibit Viral Infectivity and the Interaction with HIV.J Immunol Res. 2017;2017:1548905. doi: 10.1155/2017/1548905. Epub 2017 Nov 22. J Immunol Res. 2017. PMID: 29359168 Free PMC article. Review.
Cited by
-
The KT Jeang retrovirology prize 2024: Walther Mothes.Retrovirology. 2024 Oct 24;21(1):16. doi: 10.1186/s12977-024-00649-8. Retrovirology. 2024. PMID: 39449025 Free PMC article. No abstract available.
-
A cholesterol switch controls phospholipid scrambling by G protein-coupled receptors.J Biol Chem. 2024 Feb;300(2):105649. doi: 10.1016/j.jbc.2024.105649. Epub 2024 Jan 16. J Biol Chem. 2024. PMID: 38237683 Free PMC article.
-
Interactions between HIV proteins and host restriction factors: implications for potential therapeutic intervention in HIV infection.Front Immunol. 2024 Aug 16;15:1390650. doi: 10.3389/fimmu.2024.1390650. eCollection 2024. Front Immunol. 2024. PMID: 39221250 Free PMC article. Review.
-
New Insights into HIV Life Cycle, Th1/Th2 Shift during HIV Infection and Preferential Virus Infection of Th2 Cells: Implications of Early HIV Treatment Initiation and Care.Life (Basel). 2024 Jan 9;14(1):104. doi: 10.3390/life14010104. Life (Basel). 2024. PMID: 38255719 Free PMC article. Review.
-
Beyond Impairment of Virion Infectivity: New Activities of the Anti-HIV Host Cell Factor SERINC5.Viruses. 2024 Feb 12;16(2):284. doi: 10.3390/v16020284. Viruses. 2024. PMID: 38400059 Free PMC article. Review.
References
-
- Inuzuka M, Hayakawa M, Ingi T. Serinc, an activity-regulated protein family, incorporates serine into membrane lipid synthesis. J. Biol. Chem. 2005;280:35776–35783. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

