The disordered C terminus of ALKBH5 promotes phase separation and paraspeckles assembly
- PMID: 37474102
- PMCID: PMC10457456
- DOI: 10.1016/j.jbc.2023.105071
The disordered C terminus of ALKBH5 promotes phase separation and paraspeckles assembly
Abstract
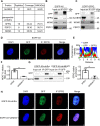
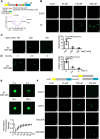
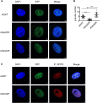
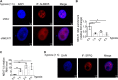
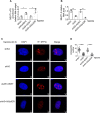
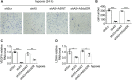
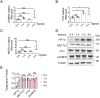
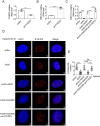
Paraspeckles (PS) are nuclear structures scaffolded by the long noncoding RNA NEAT1 and protein components such as NONO and SFPQ. We previously found that the upregulation of RNA N6-methyl-adenosine (m6A) demethylase ALKBH5 facilitates hypoxia-induced paraspeckle assembly through erasing m6A marks on NEAT1, thus stabilizing it. However, it remains unclear how these processes are spatiotemporally coordinated. Here we discover that ALKBH5 specifically binds to proteins in PS and forms phase-separated droplets that are incorporated into PS through its C-terminal intrinsically disordered region (cIDR). Upon exposure to hypoxia, rapid ALKBH5 condensation in PS induces m6A demethylation of NEAT1, which further facilitates PS formation before the upregulation of ALKBH5 expression. In cells expressing ALKBH5 lacking cIDR, PS fail to be formed in response to hypoxia, accompanied with insufficient m6A demethylation of NEAT1 and its destabilization. We also demonstrate that ALKBH5-cIDR is indispensable for hypoxia-induced effects such as cancer cell invasion. Therefore, our study has identified the role of ALKBH5 in phase separation as the molecular basis of the positive feedback loop for PS formation between ALKBH5 incorporation into PS and NEAT1 stabilization.
Keywords: ALKBH5; NEAT1; m(6)A; paraspeckle; phase separation.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures

Similar articles
-
ALKBH5 Facilitates Hypoxia-Induced Paraspeckle Assembly and IL8 Secretion to Generate an Immunosuppressive Tumor Microenvironment.Cancer Res. 2021 Dec 1;81(23):5876-5888. doi: 10.1158/0008-5472.CAN-21-1456. Epub 2021 Oct 20. Cancer Res. 2021. PMID: 34670781
-
ALKBH5 promotes invasion and metastasis of gastric cancer by decreasing methylation of the lncRNA NEAT1.J Physiol Biochem. 2019 Aug;75(3):379-389. doi: 10.1007/s13105-019-00690-8. Epub 2019 Jul 9. J Physiol Biochem. 2019. PMID: 31290116 Free PMC article.
-
ALKBH5 promotes the progression of infantile hemangioma through regulating the NEAT1/miR-378b/FOSL1 axis.Mol Cell Biochem. 2022 May;477(5):1527-1540. doi: 10.1007/s11010-022-04388-2. Epub 2022 Feb 18. Mol Cell Biochem. 2022. PMID: 35182329
-
Molecular anatomy of the architectural NEAT1 noncoding RNA: The domains, interactors, and biogenesis pathway required to build phase-separated nuclear paraspeckles.Wiley Interdiscip Rev RNA. 2019 Nov;10(6):e1545. doi: 10.1002/wrna.1545. Epub 2019 May 1. Wiley Interdiscip Rev RNA. 2019. PMID: 31044562 Review.
-
Organization and function of paraspeckles.Essays Biochem. 2020 Dec 7;64(6):875-882. doi: 10.1042/EBC20200010. Essays Biochem. 2020. PMID: 32830222 Review.
Cited by
-
The role of m6A-associated membraneless organelles in the RNA metabolism processes and human diseases.Theranostics. 2024 Aug 6;14(12):4683-4700. doi: 10.7150/thno.99019. eCollection 2024. Theranostics. 2024. PMID: 39239525 Free PMC article. Review.
-
Liquid-liquid phase separation in diseases.MedComm (2020). 2024 Jul 13;5(7):e640. doi: 10.1002/mco2.640. eCollection 2024 Jul. MedComm (2020). 2024. PMID: 39006762 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

