Conformational exchange at a C2H2 zinc-binding site facilitates redox sensing by the PML protein
- PMID: 37473756
- PMCID: PMC10528520
- DOI: 10.1016/j.str.2023.06.014
Conformational exchange at a C2H2 zinc-binding site facilitates redox sensing by the PML protein
Abstract
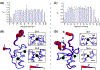
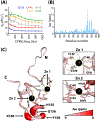
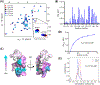
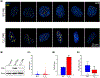
The promyelocytic leukemia protein, PML, plays a vital role in the cellular response to oxidative stress; however, the molecular mechanism of its action remains poorly understood. Here, we identify redox-sensitive sites of PML. A molecule of PML is cysteine-rich and contains three zinc-binding domains including RING, B-box1, and B-box2. Using in vitro assays, we have compared the sensitivity of the isolated RING and B-box1 domains and shown that B-box1 is more sensitive to oxidation. NMR studies of PML dynamics showed that one of the Zn-coordination sites within the B-box1 undergoes significant conformational exchange, revealing a hotspot for exposure of reactive cysteines. In agreement with the in vitro data, enhancement of the B-box1 Zn-coordination dynamics led to more efficient recruitment of PML into PML nuclear bodies in cells. Overall, our results suggest that the increased sensitivity of B-box1 to oxidative stress makes this domain an important redox-sensing component of PML.
Keywords: Carr-Purcell-Meiboom-Gill (CPMG); PML nuclear bodies; conformational change; molecular dynamics; nuclear magnetic resonance (NMR); oxidative stress; promyelocytic leukemia (PML); relaxation dispersion; zinc finger.
Copyright © 2023 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare that they have no conflicts of interest with the contents of this article.
Figures



Similar articles
-
The solution structure of the RING finger domain from the acute promyelocytic leukaemia proto-oncoprotein PML.EMBO J. 1995 Apr 3;14(7):1532-41. doi: 10.1002/j.1460-2075.1995.tb07139.x. EMBO J. 1995. PMID: 7729428 Free PMC article.
-
Promyelocytic Leukemia Protein, a Protein at the Crossroad of Oxidative Stress and Metabolism.Antioxid Redox Signal. 2017 Mar 20;26(9):432-444. doi: 10.1089/ars.2016.6898. Epub 2016 Dec 12. Antioxid Redox Signal. 2017. PMID: 27758112 Review.
-
PML-Nuclear Bodies Regulate the Stability of the Fusion Protein Dendra2-Nrf2 in the Nucleus.Cell Physiol Biochem. 2018;47(2):800-816. doi: 10.1159/000490033. Epub 2018 May 22. Cell Physiol Biochem. 2018. PMID: 29807365 Free PMC article.
-
In vivo and in vitro characterization of the B1 and B2 zinc-binding domains from the acute promyelocytic leukemia protooncoprotein PML.Proc Natl Acad Sci U S A. 1996 Feb 20;93(4):1601-6. doi: 10.1073/pnas.93.4.1601. Proc Natl Acad Sci U S A. 1996. PMID: 8643677 Free PMC article.
-
PML Nuclear bodies: the cancer connection and beyond.Nucleus. 2024 Dec;15(1):2321265. doi: 10.1080/19491034.2024.2321265. Epub 2024 Feb 27. Nucleus. 2024. PMID: 38411156 Free PMC article. Review.
Cited by
-
Deletions of conserved extracytoplasmic function sigma factors-encoding genes in Streptomyces have a major impact on secondary metabolism.Microb Cell Fact. 2024 Jul 18;23(1):201. doi: 10.1186/s12934-024-02479-x. Microb Cell Fact. 2024. PMID: 39026318 Free PMC article.
References
-
- Lallemand-Breitenbach V, Zhu J, Puvion F, Koken M, Honoré N, Doubeikovsky A, Duprez E, Pandolfi PP, Puvion E, Freemont P, and de Thé H (2001). Role of promyelocytic leukemia (PML) sumolation in nuclear body formation, 11S proteasome recruitment, and As2O3 - induced PML or PML/retinoic acid receptor alpha degradation. The Journal of experimental medicine 193, 1361–1371. 10.1084/JEM.193.12.1361. - DOI - PMC - PubMed
-
- Luciani JJ, Depetris D, Usson Y, Metzler-Guillemain C, Mignon-Ravix C, Mitchell MJ, Megarbane A, Sarda P, Sirma H, Moncla A, et al. (2006). PML nuclear bodies are highly organised DNA-protein structures with a function in heterochromatin remodelling at the G2 phase. Journal of cell science 119, 2518–2531. 10.1242/jcs.02965. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

