Study of the regulatory elements of the Ovalbumin gene promoter using CRISPR technology in chicken cells
- PMID: 37461059
- PMCID: PMC10353141
- DOI: 10.1186/s13036-023-00367-3
Study of the regulatory elements of the Ovalbumin gene promoter using CRISPR technology in chicken cells
Abstract
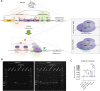
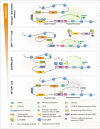
Background: Hormone-dependent promoters are very efficient in transgene expression. Plasmid-based reporter assays have identified regulatory sequences of the Ovalbumin promoter that are involved in response to estrogen and have shown that the deletion of the steroid-dependent regulatory element (SDRE) and negative regulatory element (NRE) leads to a steroid-independent expression of a reporter. However, the functional roles of these regulatory elements within the native genomic context of the Ovalbumin promoter have not been evaluated.
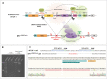
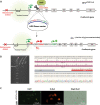
Results: In this study, we show that the negative effects of the NRE element on the Ovalbumin gene can be counteracted by CRISPR interference. We also show that the CRISPR-mediated deletion of SDRE and NRE promoter elements in a non-oviduct cell can lead to the significant expression of the Ovalbumin gene. In addition, the targeted knock-in of a transgene reporter in the Ovalbumin coding region and its expression confirms that the truncated promoter of the Ovalbumin gene can be efficiently used for an estrogen-independent expression of a foreign gene.
Conclusions: The methodology applied in this paper allowed the study of promoter regulatory sequences in their native nuclear organization.
Keywords: Avian expression systems; CRISPR technology; Chicken fibroblast; Gene editing; Ovalbumin promoter; Regulatory sequences.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
The steroid-dependent regulatory element in the ovalbumin gene does not function as a typical steroid response element.J Biol Chem. 1990 May 5;265(13):7590-5. J Biol Chem. 1990. PMID: 2332444
-
Repression of the ovalbumin gene involves multiple negative elements including a ubiquitous transcriptional silencer.Mol Endocrinol. 1995 Sep;9(9):1113-26. doi: 10.1210/mend.9.9.7491104. Mol Endocrinol. 1995. PMID: 7491104
-
Modulation of transcriptional activity of the chicken ovalbumin gene promoter in primary cultures of chicken oviduct cells: effects of putative regulatory elements in the 5'-flanking region.Biochem Mol Biol Int. 1995 Jul;36(4):811-6. Biochem Mol Biol Int. 1995. PMID: 8528143
-
A nuclear matrix acceptor site for the progesterone receptor in the avian c-myc gene promoter.Recent Prog Horm Res. 1996;51:63-96. Recent Prog Horm Res. 1996. PMID: 8701093 Review.
-
Beyond knockouts: fine-tuning regulation of gene expression in plants with CRISPR-Cas-based promoter editing.New Phytol. 2023 Aug;239(3):868-874. doi: 10.1111/nph.19020. Epub 2023 Jun 6. New Phytol. 2023. PMID: 37282668 Review.
Cited by
-
Exploring the Function of Gene Promoter Regulatory Elements Using CRISPR Tools.Methods Mol Biol. 2024;2844:145-156. doi: 10.1007/978-1-0716-4063-0_10. Methods Mol Biol. 2024. PMID: 39068338
-
Comparison of Multiple Strategies for Precision Transgene Knock-In in Gallus gallus Genome via Microhomology-Mediated End Joining.Int J Mol Sci. 2023 Oct 29;24(21):15731. doi: 10.3390/ijms242115731. Int J Mol Sci. 2023. PMID: 37958714 Free PMC article.
References
LinkOut - more resources
Full Text Sources

