mRNA lipid nanoparticle-mediated pyroptosis sensitizes immunologically cold tumors to checkpoint immunotherapy
- PMID: 37454146
- PMCID: PMC10349854
- DOI: 10.1038/s41467-023-39938-9
mRNA lipid nanoparticle-mediated pyroptosis sensitizes immunologically cold tumors to checkpoint immunotherapy
Abstract
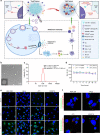
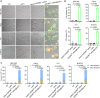
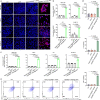
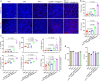
Synergistically improving T-cell responsiveness is promising for favorable therapeutic outcomes in immunologically cold tumors, yet current treatments often fail to induce a cascade of cancer-immunity cycle for effective antitumor immunity. Gasdermin-mediated pyroptosis is a newly discovered mechanism in cancer immunotherapy; however, cleavage in the N terminus is required to activate pyroptosis. Here, we report a single-agent mRNA nanomedicine-based strategy that utilizes mRNA lipid nanoparticles (LNPs) encoding only the N-terminus of gasdermin to trigger pyroptosis, eliciting robust antitumor immunity. In multiple female mouse models, we show that pyroptosis-triggering mRNA/LNPs turn cold tumors into hot ones and create a positive feedback loop to promote antitumor immunity. Additionally, mRNA/LNP-induced pyroptosis sensitizes tumors to anti-PD-1 immunotherapy, facilitating tumor growth inhibition. Antitumor activity extends beyond the treated lesions and suppresses the growth of distant tumors. We implement a strategy for inducing potent antitumor immunity, enhancing immunotherapy responses in immunologically cold tumors.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
Vesicular stomatitis virus sensitizes immunologically cold tumors to checkpoint blockade by inducing pyroptosis.Biochim Biophys Acta Mol Basis Dis. 2022 Dec 1;1868(12):166538. doi: 10.1016/j.bbadis.2022.166538. Epub 2022 Sep 9. Biochim Biophys Acta Mol Basis Dis. 2022. PMID: 36096276
-
Oncolytic Parapoxvirus induces Gasdermin E-mediated pyroptosis and activates antitumor immunity.Nat Commun. 2023 Jan 14;14(1):224. doi: 10.1038/s41467-023-35917-2. Nat Commun. 2023. PMID: 36641456 Free PMC article.
-
A Mild Hyperthermia Hollow Carbon Nanozyme as Pyroptosis Inducer for Boosted Antitumor Immunity.ACS Nano. 2023 Nov 28;17(22):22844-22858. doi: 10.1021/acsnano.3c07601. Epub 2023 Nov 9. ACS Nano. 2023. PMID: 37942890
-
Pyroptosis: A road to next-generation cancer immunotherapy.Semin Immunol. 2023 Jul;68:101782. doi: 10.1016/j.smim.2023.101782. Epub 2023 Jun 9. Semin Immunol. 2023. PMID: 37302166 Review.
-
Lighting a Fire: Can We Harness Pyroptosis to Ignite Antitumor Immunity?Cancer Immunol Res. 2021 Jan;9(1):2-7. doi: 10.1158/2326-6066.CIR-20-0525. Cancer Immunol Res. 2021. PMID: 33397791 Free PMC article. Review.
Cited by
-
Nanoparticle-mediated cell pyroptosis: a new therapeutic strategy for inflammatory diseases and cancer.J Nanobiotechnology. 2024 Aug 22;22(1):504. doi: 10.1186/s12951-024-02763-3. J Nanobiotechnology. 2024. PMID: 39175020 Free PMC article. Review.
-
A new era of cancer immunotherapy: combining revolutionary technologies for enhanced CAR-M therapy.Mol Cancer. 2024 Jun 1;23(1):117. doi: 10.1186/s12943-024-02032-9. Mol Cancer. 2024. PMID: 38824567 Free PMC article. Review.
-
Recent Advancements in mRNA Vaccines: From Target Selection to Delivery Systems.Vaccines (Basel). 2024 Aug 1;12(8):873. doi: 10.3390/vaccines12080873. Vaccines (Basel). 2024. PMID: 39203999 Free PMC article. Review.
-
Bendamustine-rituximab elicits dual tumoricidal and immunomodulatory responses via cGAS-STING activation in diffuse large B-cell lymphoma.J Immunother Cancer. 2024 Nov 9;12(11):e009212. doi: 10.1136/jitc-2024-009212. J Immunother Cancer. 2024. PMID: 39521616 Free PMC article.
-
Editorial: Advances of novel approaches to enhance therapeutic efficacy and safety in human solid cold tumor.Front Immunol. 2024 Mar 22;15:1398270. doi: 10.3389/fimmu.2024.1398270. eCollection 2024. Front Immunol. 2024. PMID: 38585262 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

