HIV-1 promotes ubiquitination of the amyloidogenic C-terminal fragment of APP to support viral replication
- PMID: 37454116
- PMCID: PMC10349857
- DOI: 10.1038/s41467-023-40000-x
HIV-1 promotes ubiquitination of the amyloidogenic C-terminal fragment of APP to support viral replication
Abstract
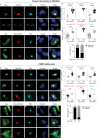
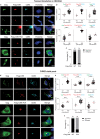
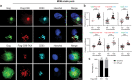
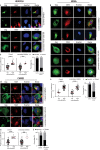
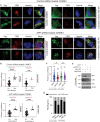
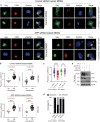
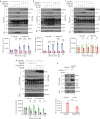
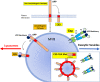
HIV-1 replication in macrophages and microglia involves intracellular assembly and budding into modified subsets of multivesicular bodies (MVBs), which support both viral persistence and spread. However, the cellular factors that regulate HIV-1's vesicular replication remain poorly understood. Recently, amyloid precursor protein (APP) was identified as an inhibitor of HIV-1 replication in macrophages and microglia via an unknown mechanism. Here, we show that entry of HIV-1 Gag into MVBs is blocked by the amyloidogenic C-terminal fragment of APP, "C99", but not by the non-amyloidogenic product, "C83". To counter this, Gag promotes multi-site ubiquitination of C99 which controls both exocytic sorting of MVBs and further processing of C99 into toxic amyloids. Processing of C99, entry of Gag into MVBs and release of infectious virus could be suppressed by expressing ubiquitination-defective C99 or by γ-secretase inhibitor treatment, suggesting that APP's amyloidogenic pathway functions to sense and suppress HIV-1 replication in macrophages and microglia.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
The A673T mutation in the amyloid precursor protein reduces the production of β-amyloid protein from its β-carboxyl terminal fragment in cells.Acta Neuropathol Commun. 2015 Nov 4;3:66. doi: 10.1186/s40478-015-0247-6. Acta Neuropathol Commun. 2015. PMID: 26531305 Free PMC article.
-
The C99 domain of the amyloid precursor protein resides in the disordered membrane phase.J Biol Chem. 2021 Jan-Jun;296:100652. doi: 10.1016/j.jbc.2021.100652. Epub 2021 Apr 9. J Biol Chem. 2021. PMID: 33839158 Free PMC article.
-
Turnover of C99 is controlled by a crosstalk between ERAD and ubiquitin-independent lysosomal degradation in human neuroglioma cells.PLoS One. 2013 Dec 20;8(12):e83096. doi: 10.1371/journal.pone.0083096. eCollection 2013. PLoS One. 2013. PMID: 24376644 Free PMC article.
-
Is γ-secretase a beneficial inactivating enzyme of the toxic APP C-terminal fragment C99?J Biol Chem. 2021 Jan-Jun;296:100489. doi: 10.1016/j.jbc.2021.100489. Epub 2021 Mar 1. J Biol Chem. 2021. PMID: 33662398 Free PMC article. Review.
-
Amyloidogenic and anti-amyloidogenic properties of presenilin 1.Adv Pharmacol. 2021;90:239-251. doi: 10.1016/bs.apha.2020.09.010. Epub 2020 Oct 24. Adv Pharmacol. 2021. PMID: 33706935 Review.
Cited by
-
Release of P-TEFb from the Super Elongation Complex promotes HIV-1 latency reversal.PLoS Pathog. 2024 Sep 11;20(9):e1012083. doi: 10.1371/journal.ppat.1012083. eCollection 2024 Sep. PLoS Pathog. 2024. PMID: 39259751 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

