The HDAC10 instructs macrophage M2 program via deacetylation of STAT3 and promotes allergic airway inflammation
- PMID: 37441601
- PMCID: PMC10334828
- DOI: 10.7150/thno.82535
The HDAC10 instructs macrophage M2 program via deacetylation of STAT3 and promotes allergic airway inflammation
Abstract
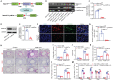
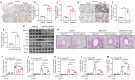
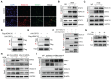
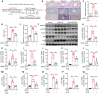
Background: Perturbation of macrophage homeostasis is one of the key mechanisms of airway inflammation in asthma. However, the exact mechanisms remain poorly understood. Objectives: We sought to examine the role of histone deacetylase (HDAC) 10 as an epigenetic regulator that governs macrophage M2 program and promotes airway inflammation in asthma, and to elucidate the underlying mechanisms. Methods: Peripheral blood and airway biopsies were obtained from healthy individuals and asthmatic patients. Asthma was induced by exposure to allergen in mice with myeloid-specific deletion of Hdac10 (Hdac10fl/fl-LysMCre) mice. HDAC10 inhibitor Salvianolic acid B (SAB), STAT3 selective agonist Colivelin, and the specific PI3K/Akt activator 1,3-Dicaffeoylquinic acid (DA) were also used in asthmatic mice. For cell studies, THP1 cells, primary mouse bone marrow derived macrophage (BMDMs) were used and related signaling pathways was investigated. Results: HDAC10 expression was highly expressed by macrophages and promoted M2 macrophage activation and airway inflammation in asthmatic patients and mice. Hdac10fl/fl-LysMCre mice were protected from airway inflammation in experimental asthma model. Hdac10 deficiency significantly attenuated STAT3 expression and decreased M2 macrophage polarization following allergen exposure. Mechanistically, HDAC10 directly binds STAT3 for deacetylation in macrophages, by which it promotes STAT3 expression and activates the macrophage M2 program. Importantly, we identified SAB as a HDAC10 inhibitor that had protective effects against airway inflammation in mice. Conclusions: Our results revealed that HDAC10-STAT3 interaction governs macrophage polarization to promote airway inflammation in asthma, implicating HDAC10 as a therapeutic target.
Keywords: HDAC10; STAT3; airway inflammation; asthma; macrophage..
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures




Similar articles
-
Myeloid-specific Hdac10 deletion protects against LPS-induced acute lung injury via P62 acetylation at lysine 165.Respir Res. 2024 Jul 2;25(1):263. doi: 10.1186/s12931-024-02891-2. Respir Res. 2024. PMID: 38956592 Free PMC article.
-
Myeloid-specific SIRT1 deletion exacerbates airway inflammatory response in a mouse model of allergic asthma.Aging (Albany NY). 2021 Jun 7;13(11):15479-15490. doi: 10.18632/aging.203104. Epub 2021 Jun 7. Aging (Albany NY). 2021. PMID: 34099590 Free PMC article.
-
FoxO1 is a critical regulator of M2-like macrophage activation in allergic asthma.Allergy. 2019 Mar;74(3):535-548. doi: 10.1111/all.13626. Epub 2018 Nov 5. Allergy. 2019. PMID: 30288751 Free PMC article.
-
Macrophages: The Good, the Bad, and the Gluttony.Front Immunol. 2021 Aug 12;12:708186. doi: 10.3389/fimmu.2021.708186. eCollection 2021. Front Immunol. 2021. PMID: 34456917 Free PMC article. Review.
-
Macrophage polarization and allergic asthma.Transl Res. 2018 Jan;191:1-14. doi: 10.1016/j.trsl.2017.09.002. Epub 2017 Oct 7. Transl Res. 2018. PMID: 29066321 Free PMC article. Review.
Cited by
-
Interplay between acetylation and ubiquitination controls PSAT1 protein stability in lung adenocarcinoma.Commun Biol. 2024 Oct 21;7(1):1365. doi: 10.1038/s42003-024-07051-2. Commun Biol. 2024. PMID: 39433916 Free PMC article.
-
Histone Deacetylase Inhibitors for Peripheral T-Cell Lymphomas.Cancers (Basel). 2024 Sep 30;16(19):3359. doi: 10.3390/cancers16193359. Cancers (Basel). 2024. PMID: 39409979 Free PMC article. Review.
-
Macrophage polarization and its impact on idiopathic pulmonary fibrosis.Front Immunol. 2024 Jul 26;15:1444964. doi: 10.3389/fimmu.2024.1444964. eCollection 2024. Front Immunol. 2024. PMID: 39131154 Free PMC article. Review.
-
Macrophage plasticity: signaling pathways, tissue repair, and regeneration.MedComm (2020). 2024 Aug 1;5(8):e658. doi: 10.1002/mco2.658. eCollection 2024 Aug. MedComm (2020). 2024. PMID: 39092292 Free PMC article. Review.
-
S100A8/9 modulates perturbation and glycolysis of macrophages in allergic asthma mice.PeerJ. 2024 Apr 18;12:e17106. doi: 10.7717/peerj.17106. eCollection 2024. PeerJ. 2024. PMID: 38646478 Free PMC article.
References
-
- Hammad H, Lambrecht BN. The basic immunology of asthma. Cell. 2021;184:1469–85. - PubMed
-
- Joshi N, Walter JM, Misharin AV. Alveolar Macrophages. Cell Immunol. 2018;330:86–90. - PubMed
-
- Sharma N, Akkoyunlu M, Rabin RL. Macrophages-common culprit in obesity and asthma. Allergy. 2018;73:1196–205. - PubMed
-
- Lechner A, Henkel FDR, Hartung F, Bohnacker S, Alessandrini F, Gubernatorova EO. et al. Macrophages acquire a TNF-dependent inflammatory memory in allergic asthma. J Allergy Clin Immunol. 2022;149:2078–90. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

