Changes in SUMO-modified proteins in Epstein-Barr virus infection identifies reciprocal regulation of TRIM24/28/33 complexes and the lytic switch BZLF1
- PMID: 37410772
- PMCID: PMC10353822
- DOI: 10.1371/journal.ppat.1011477
Changes in SUMO-modified proteins in Epstein-Barr virus infection identifies reciprocal regulation of TRIM24/28/33 complexes and the lytic switch BZLF1
Abstract
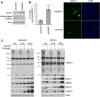
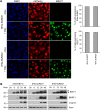
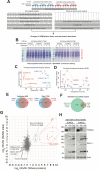
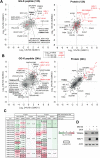
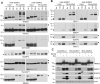
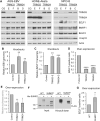
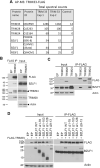
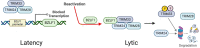
SUMO modifications regulate the function of many proteins and are important in controlling herpesvirus infections. We performed a site-specific proteomic analysis of SUMO1- and SUMO2-modified proteins in Epstein-Barr virus (EBV) latent and lytic infection to identify proteins that change in SUMO modification status in response to EBV reactivation. Major changes were identified in all three components of the TRIM24/TRIM28/TRIM33 complex, with TRIM24 being rapidly degraded and TRIM33 being phosphorylated and SUMOylated in response to EBV lytic infection. Further experiments revealed TRIM24 and TRIM33 repress expression of the EBV BZLF1 lytic switch gene, suppressing EBV reactivation. However, BZLF1 was shown to interact with TRIM24 and TRIM33, resulting in disruption of TRIM24/TRIM28/TRIM33 complexes, degradation of TRIM24 and modification followed by degradation of TRIM33. Therefore, we have identified TRIM24 and TRIM33 as cellular antiviral defence factors against EBV lytic infection and established the mechanism by which BZLF1 disables this defence.
Copyright: © 2023 De La Cruz-Herrera et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
The B-cell specific transcription factor, Oct-2, promotes Epstein-Barr virus latency by inhibiting the viral immediate-early protein, BZLF1.PLoS Pathog. 2012 Feb;8(2):e1002516. doi: 10.1371/journal.ppat.1002516. Epub 2012 Feb 9. PLoS Pathog. 2012. PMID: 22346751 Free PMC article.
-
Identification of ARKL1 as a Negative Regulator of Epstein-Barr Virus Reactivation.J Virol. 2019 Sep 30;93(20):e00989-19. doi: 10.1128/JVI.00989-19. Print 2019 Oct 15. J Virol. 2019. PMID: 31341047 Free PMC article.
-
Sumoylation of the Epstein-Barr virus BZLF1 protein inhibits its transcriptional activity and is regulated by the virus-encoded protein kinase.J Virol. 2010 May;84(9):4383-94. doi: 10.1128/JVI.02369-09. Epub 2010 Feb 24. J Virol. 2010. PMID: 20181712 Free PMC article.
-
Molecular Basis of Epstein-Barr Virus Latency Establishment and Lytic Reactivation.Viruses. 2021 Nov 23;13(12):2344. doi: 10.3390/v13122344. Viruses. 2021. PMID: 34960613 Free PMC article. Review.
-
Switching of EBV cycles between latent and lytic states.Rev Med Virol. 2014 May;24(3):142-53. doi: 10.1002/rmv.1780. Epub 2013 Dec 11. Rev Med Virol. 2014. PMID: 24339346 Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

