How host ER membrane chaperones and morphogenic proteins support virus infection
- PMID: 37401530
- PMCID: PMC10357032
- DOI: 10.1242/jcs.261121
How host ER membrane chaperones and morphogenic proteins support virus infection
Abstract
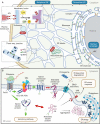
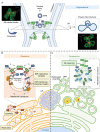
The multi-functional endoplasmic reticulum (ER) is exploited by viruses to cause infection. Morphologically, this organelle is a highly interconnected membranous network consisting of sheets and tubules whose levels are dynamic, changing in response to cellular conditions. Functionally, the ER is responsible for protein synthesis, folding, secretion and degradation, as well as Ca2+ homeostasis and lipid biosynthesis, with each event catalyzed by defined ER factors. Strikingly, these ER host factors are hijacked by viruses to support different infection steps, including entry, translation, replication, assembly and egress. Although the full repertoire of these ER factors that are hijacked is unknown, recent studies have uncovered several ER membrane machineries that are exploited by viruses - ranging from polyomavirus to flavivirus and coronavirus - to facilitate different steps of their life cycle. These discoveries should provide better understanding of virus infection mechanisms, potentially leading to the development of more effective anti-viral therapies.
Keywords: Coronavirus; ER membrane complex; ER morphogenesis; Endoplasmic reticulum; Flavivirus; Polyomavirus.
© 2023. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests B.T. is collaborating with Via Nova Therapeutics. The other authors declare no competing or financial interests.
Figures

Similar articles
-
ER functions are exploited by viruses to support distinct stages of their life cycle.Biochem Soc Trans. 2020 Oct 30;48(5):2173-2184. doi: 10.1042/BST20200395. Biochem Soc Trans. 2020. PMID: 33119046 Free PMC article. Review.
-
How DNA and RNA Viruses Exploit Host Chaperones to Promote Infection.Viruses. 2021 May 21;13(6):958. doi: 10.3390/v13060958. Viruses. 2021. PMID: 34064125 Free PMC article. Review.
-
How viruses use the endoplasmic reticulum for entry, replication, and assembly.Cold Spring Harb Perspect Biol. 2013 Jan 1;5(1):a013250. doi: 10.1101/cshperspect.a013250. Cold Spring Harb Perspect Biol. 2013. PMID: 23284050 Free PMC article. Review.
-
Endoplasmic Reticulum Chaperones in Viral Infection: Therapeutic Perspectives.Microbiol Mol Biol Rev. 2021 Dec 15;85(4):e0003521. doi: 10.1128/MMBR.00035-21. Epub 2021 Oct 13. Microbiol Mol Biol Rev. 2021. PMID: 34643441 Free PMC article. Review.
-
Membrane-Associated Flavivirus Replication Complex-Its Organization and Regulation.Viruses. 2021 Jun 3;13(6):1060. doi: 10.3390/v13061060. Viruses. 2021. PMID: 34205058 Free PMC article. Review.
Cited by
-
TTC17 is an endoplasmic reticulum resident TPR-containing adaptor protein.J Biol Chem. 2023 Dec;299(12):105450. doi: 10.1016/j.jbc.2023.105450. Epub 2023 Nov 8. J Biol Chem. 2023. PMID: 37949225 Free PMC article.
-
Prostaglandin A3 regulates the colony development of Odontotermes formosanus by reducing worker proportion.Crop Health. 2024;2(1):11. doi: 10.1007/s44297-024-00030-3. Epub 2024 Jul 2. Crop Health. 2024. PMID: 38984319 Free PMC article.
-
Host mitochondria: more than an organelle in SARS-CoV-2 infection.Front Cell Infect Microbiol. 2023 Aug 25;13:1228275. doi: 10.3389/fcimb.2023.1228275. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37692170 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

