This is a preprint.
HIV-1 latency reversal agent boosting is not limited by opioid use
- PMID: 37398278
- PMCID: PMC10312897
- DOI: 10.1101/2023.05.26.23290576
HIV-1 latency reversal agent boosting is not limited by opioid use
Update in
-
HIV-1 latency reversal agent boosting is not limited by opioid use.JCI Insight. 2024 Nov 22;9(22):e185480. doi: 10.1172/jci.insight.185480. JCI Insight. 2024. PMID: 39470739 Free PMC article.
Abstract
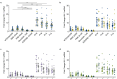
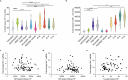
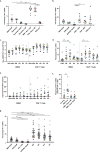
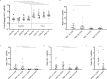
The opioid epidemic may impact the HIV-1 reservoir and its reversal from latency in virally suppressed people with HIV (PWH). We studied forty-seven PWH and observed that lowering the concentration of HIV-1 latency reversal agents (LRA), used in combination with small molecules that do not reverse latency, synergistically increases the magnitude of HIV-1 re-activation ex vivo, regardless of opioid use. This LRA boosting, which combines a Smac mimetic or low-dose protein kinase C agonist with histone deacetylase inhibitors, can generate significantly more unspliced HIV-1 transcription than phorbol 12-myristate 13-acetate (PMA) with ionomycin (PMAi), the maximal known HIV-1 reactivator. LRA boosting associated with greater histone acetylation in CD4+ T cells and modulated T cell activation-induced markers and intracellular cytokine production; Smac mimetic-based boosting was less likely to induce immune activation. We found that HIV-1 reservoirs in PWH contain unspliced and polyadenylated (polyA) virus mRNA, the ratios of which are greater in resting than total CD4+ T cells and can correct to 1:1 with PMAi exposure. Latency reversal results in greater fold-change increases to HIV-1 poly(A) mRNA than unspliced message. Multiply spliced HIV-1 transcripts and virion production did not consistently increase with LRA boosting, suggesting the presence of a persistent post-transcriptional block. LRA boosting can be leveraged to probe the mechanisms of an effective cellular HIV-1 latency reversal program.
Figures


Similar articles
-
HIV-1 latency reversal agent boosting is not limited by opioid use.JCI Insight. 2024 Nov 22;9(22):e185480. doi: 10.1172/jci.insight.185480. JCI Insight. 2024. PMID: 39470739 Free PMC article.
-
Multiply spliced HIV RNA is a predictive measure of virus production ex vivo and in vivo following reversal of HIV latency.EBioMedicine. 2021 Mar;65:103241. doi: 10.1016/j.ebiom.2021.103241. Epub 2021 Feb 26. EBioMedicine. 2021. PMID: 33647768 Free PMC article.
-
Ex vivo analysis identifies effective HIV-1 latency-reversing drug combinations.J Clin Invest. 2015 May;125(5):1901-12. doi: 10.1172/JCI80142. Epub 2015 Mar 30. J Clin Invest. 2015. PMID: 25822022 Free PMC article.
-
CXCR4 Targeting Nanoplatform for Transcriptional Activation of Latent HIV-1 Infected T Cells.ACS Appl Bio Mater. 2024 Aug 19;7(8):4831-4842. doi: 10.1021/acsabm.3c00456. Epub 2023 Aug 16. ACS Appl Bio Mater. 2024. PMID: 37586084 Review.
-
Latency Reversal 2.0: Giving the Immune System a Seat at the Table.Curr HIV/AIDS Rep. 2021 Apr;18(2):117-127. doi: 10.1007/s11904-020-00540-z. Epub 2021 Jan 12. Curr HIV/AIDS Rep. 2021. PMID: 33433817 Free PMC article. Review.
References
-
- Wong JK, Hezareh M, Gunthard HF, Havlir DV, Ignacio CC, Spina CA, et al. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science (New York, NY. 1997;278(5341):1291–5. - PubMed
-
- Finzi D, Blankson J, Siliciano JD, Margolick JB, Chadwick K, Pierson T, et al. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nature medicine. 1999;5(5):512–7. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
