Nuclear lamina strain states revealed by intermolecular force biosensor
- PMID: 37391402
- PMCID: PMC10313699
- DOI: 10.1038/s41467-023-39563-6
Nuclear lamina strain states revealed by intermolecular force biosensor
Abstract
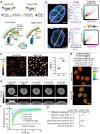
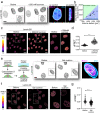
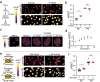
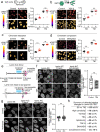
Nuclear lamins have been considered an important structural element of the nucleus. The nuclear lamina is thought both to shield DNA from excessive mechanical forces and to transmit mechanical forces onto the DNA. However, to date there is not yet a technical approach to directly measure mechanical forces on nuclear lamins at the protein level. To overcome this limitation, we developed a nanobody-based intermolecular tension FRET biosensor capable of measuring the mechanical strain of lamin filaments. Using this sensor, we were able to show that the nuclear lamina is subjected to significant force. These forces are dependent on nuclear volume, actomyosin contractility, functional LINC complex, chromatin condensation state, cell cycle, and EMT. Interestingly, large forces were also present on nucleoplasmic lamins, indicating that these lamins may also have an important mechanical role in the nucleus. Overall, we demonstrate that the nanobody-based approach allows construction of biosensors for complex protein structures for mechanobiology studies.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Scaffold, mechanics and functions of nuclear lamins.FEBS Lett. 2023 Nov;597(22):2791-2805. doi: 10.1002/1873-3468.14750. Epub 2023 Oct 21. FEBS Lett. 2023. PMID: 37813648 Review.
-
Structure and unique mechanical aspects of nuclear lamin filaments.Curr Opin Struct Biol. 2020 Oct;64:152-159. doi: 10.1016/j.sbi.2020.06.017. Epub 2020 Aug 15. Curr Opin Struct Biol. 2020. PMID: 32810798 Review.
-
Nuclear lamin isoforms differentially contribute to LINC complex-dependent nucleocytoskeletal coupling and whole-cell mechanics.Proc Natl Acad Sci U S A. 2022 Apr 26;119(17):e2121816119. doi: 10.1073/pnas.2121816119. Epub 2022 Apr 19. Proc Natl Acad Sci U S A. 2022. PMID: 35439057 Free PMC article.
-
Filaments made from A- and B-type lamins differ in structure and organization.J Cell Sci. 2008 Jan 15;121(Pt 2):215-25. doi: 10.1242/jcs.022020. J Cell Sci. 2008. PMID: 18187453
-
[Research progress in A-type lamins].Sheng Li Ke Xue Jin Zhan. 2009 Jul;40(3):203-8. Sheng Li Ke Xue Jin Zhan. 2009. PMID: 19803422 Review. Chinese.
Cited by
-
A Versatile Micromanipulation Apparatus for Biophysical Assays of the Cell Nucleus.Cell Mol Bioeng. 2022 Sep 6;15(4):303-312. doi: 10.1007/s12195-022-00734-y. eCollection 2022 Aug. Cell Mol Bioeng. 2022. PMID: 36119136 Free PMC article.
-
Matrix stiffness drives drop like nuclear deformation and lamin A/C tension-dependent YAP nuclear localization.Nat Commun. 2024 Nov 22;15(1):10151. doi: 10.1038/s41467-024-54577-4. Nat Commun. 2024. PMID: 39578439 Free PMC article.
-
Comparison of vinculin tension in cellular monolayers and three-dimensional multicellular aggregates.Biomed Opt Express. 2024 Aug 13;15(9):5199-5214. doi: 10.1364/BOE.529156. eCollection 2024 Sep 1. Biomed Opt Express. 2024. PMID: 39296399 Free PMC article.
-
Genetically Encoded Fluorescence Resonance Energy Transfer Biosensor for Live-Cell Visualization of Lamin A Phosphorylation at Serine 22.Biomater Res. 2024 Oct 22;28:0091. doi: 10.34133/bmr.0091. eCollection 2024. Biomater Res. 2024. PMID: 39440272 Free PMC article.
-
Rethinking nuclear shaping: insights from the nuclear drop model.Soft Matter. 2024 Oct 2;20(38):7558-7565. doi: 10.1039/d4sm00683f. Soft Matter. 2024. PMID: 39105242 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

