Proteomic and functional analysis of HDL subclasses in humans and rats: a proof-of-concept study
- PMID: 37386457
- PMCID: PMC10308741
- DOI: 10.1186/s12944-023-01829-9
Proteomic and functional analysis of HDL subclasses in humans and rats: a proof-of-concept study
Abstract
Background: The previous study investigated whether the functions of small, medium, and large high density lipoprotein (S/M/L-HDL) are correlated with protein changes in mice. Herein, the proteomic and functional analyses of high density lipoprotein (HDL) subclasses were performed in humans and rats.
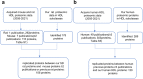
Methods: After purifying S/M/L-HDL subclasses from healthy humans (n = 6) and rats (n = 3) using fast protein liquid chromatography (FPLC) with calcium silica hydrate (CSH) resin, the proteomic analysis by mass spectrometry was conducted, as well as the capacities of cholesterol efflux and antioxidation was measured.
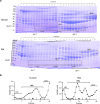
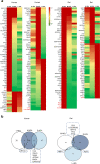
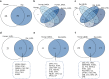
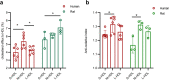
Results: Of the 120 and 106 HDL proteins identified, 85 and 68 proteins were significantly changed in concentration among the S/M/L-HDL subclasses in humans and rats, respectively. Interestingly, it was found that the relatively abundant proteins in the small HDL (S-HDL) and large HDL (L-HDL) subclasses did not overlap, both in humans and in rats. Next, by searching for the biological functions of the relatively abundant proteins in the HDL subclasses via Gene Ontology, it was displayed that the relatively abundant proteins involved in lipid metabolism and antioxidation were enriched more in the medium HDL (M-HDL) subclass than in the S/L-HDL subclasses in humans, whereas in rats, the relatively abundant proteins associated with lipid metabolism and anti-oxidation were enriched in M/L-HDL and S/M-HDL, respectively. Finally, it was confirmed that M-HDL and L-HDL had the highest cholesterol efflux capacity among the three HDL subclasses in humans and rats, respectively; moreover, M-HDL exhibited higher antioxidative capacity than S-HDL in both humans and rats.
Conclusions: The S-HDL and L-HDL subclasses are likely to have different proteomic components during HDL maturation, and results from the proteomics-based comparison of the HDL subclasses may explain the associated differences in function.
Keywords: Antioxidation; Cholesterol efflux; Fast protein liquid chromatography; HDL subclass; High density lipoprotein; Proteomics.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
HDL subclass proteomic analysis and functional implication of protein dynamic change during HDL maturation.Redox Biol. 2019 Jun;24:101222. doi: 10.1016/j.redox.2019.101222. Epub 2019 May 17. Redox Biol. 2019. PMID: 31153037 Free PMC article.
-
Serum cholesterol and triglyceride reference ranges of twenty lipoprotein subclasses for healthy Japanese men and women.Atherosclerosis. 2013 Dec;231(2):238-45. doi: 10.1016/j.atherosclerosis.2013.09.008. Epub 2013 Oct 1. Atherosclerosis. 2013. PMID: 24267234
-
Glycemic control and high-density lipoprotein characteristics in adolescents with type 1 diabetes.Pediatr Diabetes. 2013 Sep;14(6):399-406. doi: 10.1111/j.1399-5448.2012.00924.x. Epub 2012 Oct 11. Pediatr Diabetes. 2013. PMID: 23057424
-
The relationship between high density lipoprotein subclass profile and plasma lipids concentrations.Lipids Health Dis. 2010 Oct 17;9:118. doi: 10.1186/1476-511X-9-118. Lipids Health Dis. 2010. PMID: 20950490 Free PMC article. Review.
-
Proteomics of apolipoproteins and associated proteins from plasma high-density lipoproteins.Arterioscler Thromb Vasc Biol. 2010 Feb;30(2):156-63. doi: 10.1161/ATVBAHA.108.179317. Epub 2009 Sep 24. Arterioscler Thromb Vasc Biol. 2010. PMID: 19778948 Review.
Cited by
-
High-density lipoprotein functionality in cholesterol efflux in early childhood is related to the content ratio of triglyceride to cholesterol.Sci Rep. 2024 Oct 7;14(1):23323. doi: 10.1038/s41598-024-74699-5. Sci Rep. 2024. PMID: 39375444 Free PMC article.
-
Impact of a Mediterranean-Inspired Diet on Cardiovascular Disease Risk Factors: A Randomized Clinical Trial.Nutrients. 2024 Jul 26;16(15):2443. doi: 10.3390/nu16152443. Nutrients. 2024. PMID: 39125324 Free PMC article. Clinical Trial.
References
-
- Barter PJ, Caulfield M, Eriksson M, Grundy SM, Kastelein JJ, Komajda M, et al. Effects of torcetrapib in patients at high risk for coronary events. N Engl J Med. 2007;357:2109–2122. - PubMed
-
- Schwartz GG, Olsson AG, Abt M, Ballantyne CM, Barter PJ, Brumm J, et al. Effects of dalcetrapib in patients with a recent acute coronary syndrome. N Engl J Med. 2012;367:2089–2099. - PubMed
-
- Annema W, von Eckardstein A. High-density lipoproteins. Multifunctional but vulnerable protections from atherosclerosis. Circ J. 2013;77:2432–48. - PubMed
-
- Soria-Florido MT, Castañer O, Lassale C, Estruch R, Salas-Salvadó J, Martínez-González M, et al. Dysfunctional high-density lipoproteins are associated with a greater incidence of acute coronary syndrome in a population at high cardiovascular risk: a nested case-control study. Circulation. 2020;141:444–453. - PubMed
-
- Annema W, von Eckardstein A. Dysfunctional high-density lipoproteins in coronary heart disease: implications for diagnostics and therapy. Transl Res. 2016;173:30–57. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

