Large-scale data-driven and physics-based models offer insights into the relationships among the structures, dynamics, and functions of chromosomes
- PMID: 37365687
- PMCID: PMC10782906
- DOI: 10.1093/jmcb/mjad042
Large-scale data-driven and physics-based models offer insights into the relationships among the structures, dynamics, and functions of chromosomes
Abstract
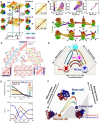
The organized three-dimensional chromosome architecture in the cell nucleus provides scaffolding for precise regulation of gene expression. When the cell changes its identity in the cell-fate decision-making process, extensive rearrangements of chromosome structures occur accompanied by large-scale adaptations of gene expression, underscoring the importance of chromosome dynamics in shaping genome function. Over the last two decades, rapid development of experimental methods has provided unprecedented data to characterize the hierarchical structures and dynamic properties of chromosomes. In parallel, these enormous data offer valuable opportunities for developing quantitative computational models. Here, we review a variety of large-scale polymer models developed to investigate the structures and dynamics of chromosomes. Different from the underlying modeling strategies, these approaches can be classified into data-driven ('top-down') and physics-based ('bottom-up') categories. We discuss their contributions to offering valuable insights into the relationships among the structures, dynamics, and functions of chromosomes and propose the perspective of developing data integration approaches from different experimental technologies and multidisciplinary theoretical/simulation methods combined with different modeling strategies.
Keywords: 4D genome; cell-fate decision; chromosome dynamics; data-driven model; physics-based model; structural changes; structure–function relationships.
© The Author(s) (2023). Published by Oxford University Press on behalf of Journal of Molecular Cell Biology, CEMCS, CAS.
Figures



Similar articles
-
Macromolecular crowding: chemistry and physics meet biology (Ascona, Switzerland, 10-14 June 2012).Phys Biol. 2013 Aug;10(4):040301. doi: 10.1088/1478-3975/10/4/040301. Epub 2013 Aug 2. Phys Biol. 2013. PMID: 23912807
-
Computational models of large-scale genome architecture.Int Rev Cell Mol Biol. 2014;307:275-349. doi: 10.1016/B978-0-12-800046-5.00009-6. Int Rev Cell Mol Biol. 2014. PMID: 24380598 Review.
-
Physics-Based Polymer Models to Probe Chromosome Structure in Single Molecules.Methods Mol Biol. 2023;2655:57-66. doi: 10.1007/978-1-0716-3143-0_5. Methods Mol Biol. 2023. PMID: 37212988
-
Simulation of Different Three-Dimensional Models of Whole Interphase Nuclei Compared to Experiments - A Consistent Scale-Bridging Simulation Framework for Genome Organization.Results Probl Cell Differ. 2022;70:495-549. doi: 10.1007/978-3-031-06573-6_18. Results Probl Cell Differ. 2022. PMID: 36348120
-
Polymer models are a versatile tool to study chromatin 3D organization.Biochem Soc Trans. 2021 Aug 27;49(4):1675-1684. doi: 10.1042/BST20201004. Biochem Soc Trans. 2021. PMID: 34282837 Review.
Cited by
-
Quantifying the large-scale chromosome structural dynamics during the mitosis-to-G1 phase transition of cell cycle.Open Biol. 2023 Nov;13(11):230175. doi: 10.1098/rsob.230175. Epub 2023 Nov 1. Open Biol. 2023. PMID: 37907089 Free PMC article.

