Metabolomic and transcriptomic analyses reveal the effects of grafting on blood orange quality
- PMID: 37360739
- PMCID: PMC10286243
- DOI: 10.3389/fpls.2023.1169220
Metabolomic and transcriptomic analyses reveal the effects of grafting on blood orange quality
Abstract
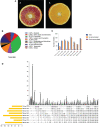
Introduction: Blood orange (Citrus sinensis L.) is a valuable source of nutrition because it is enriched in anthocyanins and has high organoleptic properties. Grafting is commonly used in citriculture and has crucial effects on various phenotypes of the blood orange, including its coloration, phenology, and biotic and abiotic resistance. Still, the underlying genetics and regulatory mechanisms are largely unexplored.
Methods: In this study, we investigated the phenotypic, metabolomic, and transcriptomic profiles at eight developmental stages of the lido blood orange cultivar (Citrus sinensis L. Osbeck cv. Lido) grafted onto two rootstocks.
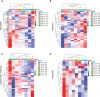
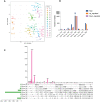
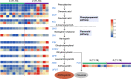
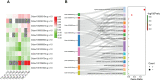
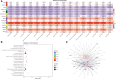
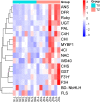
Results and discussion: The Trifoliate orange rootstock provided the best fruit quality and flesh color for Lido blood orange. Comparative metabolomics suggested significant differences in accumulation patterns of metabolites and we identified 295 differentially accumulated metabolites. The major contributors were flavonoids, phenolic acids, lignans and coumarins, and terpenoids. Moreover, transcriptome profiling resulted in the identification of 4179 differentially expressed genes (DEGs), and 54 DEGs were associated with flavonoids and anthocyanins. Weighted gene co-expression network analysis identified major genes associated to 16 anthocyanins. Furthermore, seven transcription factors (C2H2, GANT, MYB-related, AP2/ERF, NAC, bZIP, and MYB) and five genes associated with anthocyanin synthesis pathway (CHS, F3H, UFGT, and ANS) were identified as key modulators of the anthocyanin content in lido blood orange. Overall, our results revealed the impact of rootstock on the global transcriptome and metabolome in relation to fruit quality in lido blood orange. The identified key genes and metabolites can be further utilized for the quality improvement of blood orange varieties.
Keywords: blood orange; flavonoids; gene expression; pigmentation; quality improvement.
Copyright © 2023 Yang, Chen, Wang, Hou, Li, Guan, Yang, Wang and Hong.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Metabolomic and transcriptomic analyses reveal the effects of self- and hetero-grafting on anthocyanin biosynthesis in grapevine.Hortic Res. 2022 May 17;9:uhac103. doi: 10.1093/hr/uhac103. eCollection 2022. Hortic Res. 2022. PMID: 35795384 Free PMC article.
-
Weighted gene coexpression correlation network analysis reveals the potential molecular regulatory mechanism of citrate and anthocyanin accumulation between postharvest 'Bingtangcheng' and 'Tarocco' blood orange fruit.BMC Plant Biol. 2023 Jun 2;23(1):296. doi: 10.1186/s12870-023-04309-5. BMC Plant Biol. 2023. PMID: 37268922 Free PMC article.
-
Hetero-grafting affects flavonoid biosynthesis in sweet orange 'Newhall' (Citrus sinensis) peels: a metabolomics and transcriptomics analysis.Front Plant Sci. 2023 Jul 3;14:1218426. doi: 10.3389/fpls.2023.1218426. eCollection 2023. Front Plant Sci. 2023. PMID: 37465384 Free PMC article.
-
Integrated Metabolomic and Transcriptomic Analysis Reveals Differential Mechanism of Flavonoid Biosynthesis in Two Cultivars of Angelica sinensis.Molecules. 2022 Jan 4;27(1):306. doi: 10.3390/molecules27010306. Molecules. 2022. PMID: 35011537 Free PMC article.
-
The State of the Art in Biosynthesis of Anthocyanins and Its Regulation in Pigmented Sweet Oranges [(Citrus sinensis) L. Osbeck].J Agric Food Chem. 2015 Apr 29;63(16):4031-41. doi: 10.1021/acs.jafc.5b01123. Epub 2015 Apr 20. J Agric Food Chem. 2015. PMID: 25871434 Review.
Cited by
-
Combined Metabolomics and Network Pharmacology Analysis Reveal the Effect of Rootstocks on Anthocyanins, Lipids, and Potential Pharmacological Ingredients of Tarroco Blood Orange (Citrus sinensis L. Osbeck).Plants (Basel). 2024 Aug 14;13(16):2259. doi: 10.3390/plants13162259. Plants (Basel). 2024. PMID: 39204695 Free PMC article.
-
Widely Targeted Metabolomic Analysis Provides New Insights into the Effect of Rootstocks on Citrus Fruit Quality.Metabolites. 2024 Apr 21;14(4):242. doi: 10.3390/metabo14040242. Metabolites. 2024. PMID: 38668370 Free PMC article.
References
-
- Arena E., Fallico B., Maccarone E. (2001). Evaluation of antioxidant capacity of blood orange juices as influenced by constituents, concentration process and storage. Food Chem. 74, 423–427. doi: 10.1016/S0308-8146(01)00125-X - DOI
-
- Barry G. H., Castle W. S., Davies F. S. (2004). Rootstocks and plant water relations affect sugar accumulation of citrus fruit via osmotic adjustment. J. Am. Soc. Hortic. Sci. 129, 881–889. doi: 10.21273/JASHS.129.6.0881 - DOI
-
- Bowman K. D., Mccollum G., Albrecht U. (2016). Performance of ‘Valencia’orange (Citrus sinensis [L.] osbeck) on 17 rootstocks in a trial severely affected by huanglongbing. Scientia Hortic. 201, 355–361. doi: 10.1016/j.scienta.2016.01.019 - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

