Integrated multi-omics analyses reveal the altered transcriptomic characteristics of pulmonary macrophages in immunocompromised hosts with Pneumocystis pneumonia
- PMID: 37359523
- PMCID: PMC10289015
- DOI: 10.3389/fimmu.2023.1179094
Integrated multi-omics analyses reveal the altered transcriptomic characteristics of pulmonary macrophages in immunocompromised hosts with Pneumocystis pneumonia
Abstract
Introduction: With the extensive use of immunosuppressants, immunosuppression-associated pneumonitis including Pneumocystis jirovecii pneumonia (PCP) has received increasing attention. Though aberrant adaptive immunity has been considered as a key reason for opportunistic infections, the characteristics of innate immunity in these immunocompromised hosts remain unclear.
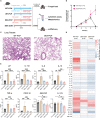
Methods: In this study, wild type C57BL/6 mice or dexamethasone-treated mice were injected with or without Pneumocystis. Bronchoalveolar lavage fluids (BALFs) were harvested for the multiplex cytokine and metabolomics analysis. The single-cell RNA sequencing (scRNA-seq) of indicated lung tissues or BALFs was performed to decipher the macrophages heterogeneity. Mice lung tissues were further analyzed via quantitative polymerase chain reaction (qPCR) or immunohistochemical staining.
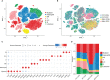
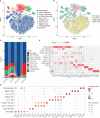
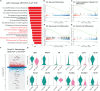
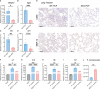
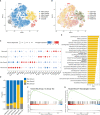
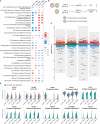
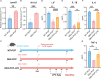
Results: We found that the secretion of both pro-inflammatory cytokines and metabolites in the Pneumocystis-infected mice are impaired by glucocorticoids. By scRNA-seq, we identified seven subpopulations of macrophages in mice lung tissues. Among them, a group of Mmp12+ macrophages is enriched in the immunocompetent mice with Pneumocystis infection. Pseudotime trajectory showed that these Mmp12+ macrophages are differentiated from Ly6c+ classical monocytes, and highly express pro-inflammatory cytokines elevated in BALFs of Pneumocystis-infected mice. In vitro, we confirmed that dexamethasone impairs the expression of Lif, Il1b, Il6 and Tnf, as well as the fungal killing capacity of alveolar macrophage (AM)-like cells. Moreover, in patients with PCP, we found a group of macrophages resembled the aforementioned Mmp12+ macrophages, and these macrophages are inhibited in the patient receiving glucocorticoid treatment. Additionally, dexamethasone simultaneously impaired the functional integrity of resident AMs and downregulated the level of lysophosphatidylcholine, leading to the suppressed antifungal capacities.
Conclusion: We reported a group of Mmp12+ macrophages conferring protection during Pneumocystis infection, which can be dampened by glucocorticoids. This study provides multiple resources for understanding the heterogeneity and metabolic changes of innate immunity in immunocompromised hosts, and also suggests that the loss of Mmp12+ macrophages population contributes to the pathogenesis of immunosuppression-associated pneumonitis.
Keywords: Pneumocystis pneumonia; glucocorticoids; immunosuppression; macrophages; single-cell RNA sequencing.
Copyright © 2023 Wang, Li, Zhao, Liu, Li, Yang, Tong and Song.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Integrated multi-omics analyses reveal the pro-inflammatory and pro-fibrotic pulmonary macrophage subcluster in silicosis.Ecotoxicol Environ Saf. 2024 Oct 1;284:116899. doi: 10.1016/j.ecoenv.2024.116899. Epub 2024 Aug 23. Ecotoxicol Environ Saf. 2024. PMID: 39181076
-
Experimental Pneumocystis lung infection promotes M2a alveolar macrophage-derived MMP12 production.Am J Physiol Lung Cell Mol Physiol. 2012 Sep;303(5):L469-75. doi: 10.1152/ajplung.00158.2012. Epub 2012 Jul 6. Am J Physiol Lung Cell Mol Physiol. 2012. PMID: 22773692 Free PMC article.
-
Intrinsic Programming of Alveolar Macrophages for Protective Antifungal Innate Immunity Against Pneumocystis Infection.Front Immunol. 2018 Sep 19;9:2131. doi: 10.3389/fimmu.2018.02131. eCollection 2018. Front Immunol. 2018. PMID: 30283457 Free PMC article.
-
Immunopathogenesis of Pneumocystis carinii pneumonia.Expert Rev Mol Med. 2005 Nov 14;7(26):1-16. doi: 10.1017/S1462399405010203. Expert Rev Mol Med. 2005. PMID: 16842636 Review.
-
Recent Advances in the Diagnosis of Pneumocystis Pneumonia.Med Mycol J. 2016;57(4):E111-E116. doi: 10.3314/mmj.16-00019. Med Mycol J. 2016. PMID: 27904052 Review.
Cited by
-
Dynamics of host immune responses and a potential function of Trem2hi interstitial macrophages in Pneumocystis pneumonia.Respir Res. 2024 Feb 5;25(1):72. doi: 10.1186/s12931-024-02709-1. Respir Res. 2024. PMID: 38317180 Free PMC article.
-
Impact of glucocorticoids and rapamycin on autophagy in Candida glabrata-infected macrophages from BALB/c mice.Front Immunol. 2024 Mar 15;15:1367048. doi: 10.3389/fimmu.2024.1367048. eCollection 2024. Front Immunol. 2024. PMID: 38585259 Free PMC article.
References
-
- Salzer HJF, Schäfer G, Hoenigl M, Günther G, Hoffmann C, Kalsdorf B, et al. . Clinical, diagnostic, and treatment disparities between hiv-infected and non-Hiv-Infected immunocompromised patients with pneumocystis jirovecii pneumonia. Respiration (2018) 96(1):52–65. doi: 10.1159/000487713 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

