KIF16B Mediates Anterograde Transport and Modulates Lysosomal Degradation of the HIV-1 Envelope Glycoprotein
- PMID: 37358446
- PMCID: PMC10373548
- DOI: 10.1128/jvi.00255-23
KIF16B Mediates Anterograde Transport and Modulates Lysosomal Degradation of the HIV-1 Envelope Glycoprotein
Abstract
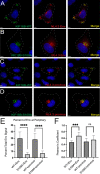
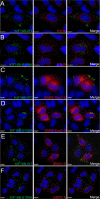
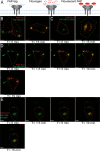
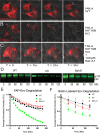
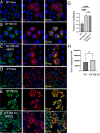
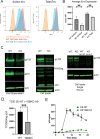
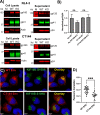
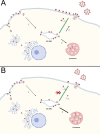
The human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein (Env) is incorporated into virions at the site of particle assembly on the plasma membrane (PM). The route taken by Env to reach the site of assembly and particle incorporation remains incompletely understood. Following initial delivery to the PM through the secretory pathway, Env is rapidly endocytosed, suggesting that recycling is required for particle incorporation. Endosomes marked by the small GTPase Rab14 have been previously shown to play a role in Env trafficking. Here, we examined the role of KIF16B, the molecular motor protein that directs outward movement of Rab14-dependent cargo, in Env trafficking. Env colocalized extensively with KIF16B+ endosomes at the cellular periphery, while expression of a motor-deficient mutant of KIF16B redistributed Env to a perinuclear location. The half-life of Env labeled at the cell surface was markedly reduced in the absence of KIF16B, while a normal half-life was restored through inhibition of lysosomal degradation. In the absence of KIF16B, Env expression on the surface of cells was reduced, leading to a reduction in Env incorporation into particles and a corresponding reduction in particle infectivity. HIV-1 replication in KIF16B knockout cells was substantially reduced compared to that in wild-type cells. These results indicated that KIF16B regulates an outward sorting step involved in Env trafficking, thereby limiting lysosomal degradation and enhancing particle incorporation. IMPORTANCE The HIV-1 envelope glycoprotein is an essential component of HIV-1 particles. The cellular pathways that contribute to incorporation of envelope into particles are not fully understood. Here, we have identified KIF16B, a motor protein that directs movement from internal compartments toward the plasma membrane, as a host factor that prevents envelope degradation and enhances particle incorporation. This is the first host motor protein identified that contributes to HIV-1 envelope incorporation and replication.
Keywords: HIV-1; KIF16B; envelope glycoprotein; kinesin; recycling; trafficking.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
HIV-1 Envelope Glycoprotein Trafficking through the Endosomal Recycling Compartment Is Required for Particle Incorporation.J Virol. 2018 Feb 12;92(5):e01893-17. doi: 10.1128/JVI.01893-17. Print 2018 Mar 1. J Virol. 2018. PMID: 29212940 Free PMC article.
-
Endocytosed HIV-1 Envelope Glycoprotein Traffics to Rab14+ Late Endosomes and Lysosomes to Regulate Surface Levels in T-Cell Lines.J Virol. 2022 Jul 27;96(14):e0076722. doi: 10.1128/jvi.00767-22. Epub 2022 Jun 30. J Virol. 2022. PMID: 35770989 Free PMC article.
-
Rab11-FIP1C and Rab14 direct plasma membrane sorting and particle incorporation of the HIV-1 envelope glycoprotein complex.PLoS Pathog. 2013;9(4):e1003278. doi: 10.1371/journal.ppat.1003278. Epub 2013 Apr 4. PLoS Pathog. 2013. PMID: 23592992 Free PMC article.
-
Viral and Host Factors Regulating HIV-1 Envelope Protein Trafficking and Particle Incorporation.Viruses. 2022 Aug 5;14(8):1729. doi: 10.3390/v14081729. Viruses. 2022. PMID: 36016351 Free PMC article. Review.
-
HIV-1 envelope glycoprotein biosynthesis, trafficking, and incorporation.J Mol Biol. 2011 Jul 22;410(4):582-608. doi: 10.1016/j.jmb.2011.04.042. J Mol Biol. 2011. PMID: 21762802 Free PMC article. Review.
Cited by
-
Tryptophan-based motifs in the LLP3 Region of the HIV-1 envelope glycoprotein cytoplasmic tail direct trafficking to the endosomal recycling compartment and mediate particle incorporation.bioRxiv [Preprint]. 2023 Apr 28:2023.04.28.538708. doi: 10.1101/2023.04.28.538708. bioRxiv. 2023. Update in: J Virol. 2023 Oct 31;97(10):e0063123. doi: 10.1128/jvi.00631-23 PMID: 37162911 Free PMC article. Updated. Preprint.
-
Second site reversion of HIV-1 envelope protein baseplate mutations maps to the matrix protein.J Virol. 2024 Feb 20;98(2):e0174223. doi: 10.1128/jvi.01742-23. Epub 2024 Jan 9. J Virol. 2024. PMID: 38193694 Free PMC article.
References
-
- Egan MA, Carruth LM, Rowell JF, Yu X, Siliciano RF. 1996. Human immunodeficiency virus type 1 envelope protein endocytosis mediated by a highly conserved intrinsic internalization signal in the cytoplasmic domain of gp41 is suppressed in the presence of the Pr55gag precursor protein. J Virol 70:6547–6556. PMC190695. doi:10.1128/JVI.70.10.6547-6556.1996. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

