TGFβ inhibition and mesenchymal to epithelial transition initiation by Xenopus egg extract: first steps towards early reprogramming in fish somatic cell
- PMID: 37339990
- PMCID: PMC10281987
- DOI: 10.1038/s41598-023-36354-3
TGFβ inhibition and mesenchymal to epithelial transition initiation by Xenopus egg extract: first steps towards early reprogramming in fish somatic cell
Abstract
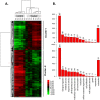
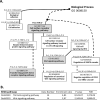
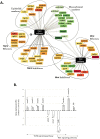
Xenopus egg extract is a powerful material to modify cultured cells fate and to induce cellular reprogramming in mammals. In this study, the response of goldfish fin cells to in vitro exposure to Xenopus egg extract, and subsequent culture, was studied using a cDNA microarray approach, gene ontology and KEGG pathways analyses, and qPCR validation. We observed that several actors of the TGFβ and Wnt/β-catenin signaling pathways, as well as some mesenchymal markers, were inhibited in treated cells, while several epithelial markers were upregulated. This was associated with morphological changes of the cells in culture, suggesting that egg extract drove cultured fin cells towards a mesenchymal-epithelial transition. This indicates that Xenopus egg extract treatment relieved some barriers of somatic reprogramming in fish cells. However, the lack of re-expression of pou2 and nanog pluripotency markers, the absence of DNA methylation remodeling of their promoter region, and the strong decrease in de novo lipid biosynthesis metabolism, indicate that reprogramming was only partial. The observed changes may render these treated cells more suitable for studies on in vivo reprogramming after somatic cell nuclear transfer.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Nuclear import of Xenopus egg extract components into cultured cells for reprogramming purposes: a case study on goldfish fin cells.Sci Rep. 2019 Feb 27;9(1):2861. doi: 10.1038/s41598-019-39500-y. Sci Rep. 2019. PMID: 30814557 Free PMC article.
-
Incomplete cellular reprogramming of colorectal cancer cells elicits an epithelial/mesenchymal hybrid phenotype.J Biomed Sci. 2018 Jul 19;25(1):57. doi: 10.1186/s12929-018-0461-1. J Biomed Sci. 2018. PMID: 30025541 Free PMC article.
-
Reprogramming of somatic cells and nuclei by Xenopus oocyte and egg extracts.Int J Dev Biol. 2016;60(7-8-9):289-296. doi: 10.1387/ijdb.160163at. Int J Dev Biol. 2016. PMID: 27251073 Review.
-
Nuclear reprogramming of human somatic cells by xenopus egg extract requires BRG1.Curr Biol. 2004 Aug 24;14(16):1475-80. doi: 10.1016/j.cub.2004.08.031. Curr Biol. 2004. PMID: 15324664
-
Reprogramming during epithelial to mesenchymal transition under the control of TGFβ.Cell Adh Migr. 2015;9(3):233-46. doi: 10.4161/19336918.2014.983794. Epub 2014 Nov 17. Cell Adh Migr. 2015. PMID: 25482613 Free PMC article. Review.
Cited by
-
Small Molecule Cocktails Promote Fibroblast-to-Leydig-like Cell Conversion for Hypogonadism Therapy.Pharmaceutics. 2023 Oct 13;15(10):2456. doi: 10.3390/pharmaceutics15102456. Pharmaceutics. 2023. PMID: 37896216 Free PMC article.
References
-
- Chenais N, Depince A, Le Bail P-Y, Labbe C. Fin cell cryopreservation and fish reconstruction by nuclear transfer stand as promising technologies for preservation of finfish genetic resources. Aquacult. Int. 2014;22:63–76. doi: 10.1007/s10499-013-9671-4. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

