Assessment and sero-diagnosis for coronaviruses with risk of human spillover
- PMID: 37334745
- PMCID: PMC10332221
- DOI: 10.1080/22221751.2023.2225932
Assessment and sero-diagnosis for coronaviruses with risk of human spillover
Abstract
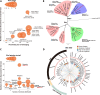
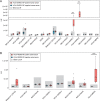
Zoonotic coronaviruses (CoVs) caused major human outbreaks in the last two decades. One of the biggest challenges during future CoV disease is ensuring rapid detection and diagnosis at the early phase of a zoonotic event, and active surveillance to the zoonotic high-risk CoVs appears the best way at the present time to provide early warnings. However, there is neither an evaluation of spillover potential nor diagnosis tools for the majority of CoVs. Here, we analyzed the viral traits, including population, genetic diversity, receptor and host species for all 40 alpha- and beta-CoV species, where the human-infecting CoVs are from. Our analysis proposed 20 high-risk CoV species, including 6 of which jumped to human, 3 with evidence of spillover but not to human and 11 without evidence of spillover yet, which prediction were further supported by an analysis of the history of CoV zoonosis. We also found three major zoonotic sources: multiple bat-origin CoV species, the rodent-origin sub-genus Embecovirus and the CoV species AlphaCoV1. Moreover, the Rhinolophidae and Hipposideridae bats harbour a significantly higher proportion of human-threatening CoV species, whereas camel, civet, swine and pangolin could be important intermediate hosts during CoV zoonotic transmission. Finally, we established quick and sensitive serologic tools for a list of proposed high-risk CoVs and validated the methods in serum cross-reaction assays using hyper-immune rabbit sera or clinical samples. By comprehensive risk assessment of the potential human-infecting CoVs, our work provides a theoretical or practical basis for future CoV disease preparedness.
Keywords: Coronavirus; bats; risk assessment; serology; spillover; zoonosis.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures


Similar articles
-
Cross-Species Transmission of Bat Coronaviruses in the Americas: Contrasting Patterns between Alphacoronavirus and Betacoronavirus.Microbiol Spectr. 2022 Aug 31;10(4):e0141122. doi: 10.1128/spectrum.01411-22. Epub 2022 Jun 30. Microbiol Spectr. 2022. PMID: 35770987 Free PMC article.
-
Coronavirus and paramyxovirus in bats from Northwest Italy.BMC Vet Res. 2017 Dec 22;13(1):396. doi: 10.1186/s12917-017-1307-x. BMC Vet Res. 2017. PMID: 29273042 Free PMC article.
-
Strategy To Assess Zoonotic Potential Reveals Low Risk Posed by SARS-Related Coronaviruses from Bat and Pangolin.mBio. 2023 Apr 25;14(2):e0328522. doi: 10.1128/mbio.03285-22. Epub 2023 Feb 14. mBio. 2023. PMID: 36786573 Free PMC article.
-
Ecology, evolution and classification of bat coronaviruses in the aftermath of SARS.Antiviral Res. 2014 Jan;101:45-56. doi: 10.1016/j.antiviral.2013.10.013. Epub 2013 Oct 31. Antiviral Res. 2014. PMID: 24184128 Free PMC article. Review.
-
Ecology, evolution and spillover of coronaviruses from bats.Nat Rev Microbiol. 2022 May;20(5):299-314. doi: 10.1038/s41579-021-00652-2. Epub 2021 Nov 19. Nat Rev Microbiol. 2022. PMID: 34799704 Free PMC article. Review.
Cited by
-
Aptamers targeting SARS-CoV-2 nucleocapsid protein exhibit potential anti pan-coronavirus activity.Signal Transduct Target Ther. 2024 Feb 14;9(1):40. doi: 10.1038/s41392-024-01748-w. Signal Transduct Target Ther. 2024. PMID: 38355661 Free PMC article.
-
UB-612 pan-SARS-CoV-2 T cell immunity-promoting vaccine protects against COVID-19 moderate-severe disease.iScience. 2024 Jan 17;27(2):108887. doi: 10.1016/j.isci.2024.108887. eCollection 2024 Feb 16. iScience. 2024. PMID: 38318376 Free PMC article.
References
-
- taxonomy I . Available from https://ictv.global/taxonomy; 2022.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

