This is a preprint.
Proteostatic tuning underpins the evolution of novel multicellular traits
- PMID: 37333256
- PMCID: PMC10274739
- DOI: 10.1101/2023.05.31.543183
Proteostatic tuning underpins the evolution of novel multicellular traits
Update in
-
Proteostatic tuning underpins the evolution of novel multicellular traits.Sci Adv. 2024 Mar 8;10(10):eadn2706. doi: 10.1126/sciadv.adn2706. Epub 2024 Mar 8. Sci Adv. 2024. PMID: 38457507 Free PMC article.
Abstract
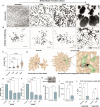
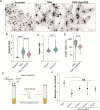
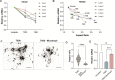
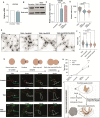
The evolution of multicellularity paved the way for the origin of complex life on Earth, but little is known about the mechanistic basis of early multicellular evolution. Here, we examine the molecular basis of multicellular adaptation in the Multicellularity Long Term Evolution Experiment (MuLTEE). We demonstrate that cellular elongation, a key adaptation underpinning increased biophysical toughness and organismal size, is convergently driven by downregulation of the chaperone Hsp90. Mechanistically, Hsp90-mediated morphogenesis operates by destabilizing the cyclin-dependent kinase Cdc28, resulting in delayed mitosis and prolonged polarized growth. Reinstatement of Hsp90 or Cdc28 expression resulted in shortened cells that formed smaller groups with reduced multicellular fitness. Together, our results show how ancient protein folding systems can be tuned to drive rapid evolution at a new level of biological individuality by revealing novel developmental phenotypes.
Conflict of interest statement
Competing interests: Authors declare that they have no competing interests.
Figures
Similar articles
-
Proteostatic tuning underpins the evolution of novel multicellular traits.Sci Adv. 2024 Mar 8;10(10):eadn2706. doi: 10.1126/sciadv.adn2706. Epub 2024 Mar 8. Sci Adv. 2024. PMID: 38457507 Free PMC article.
-
Emergence and maintenance of stable coexistence during a long-term multicellular evolution experiment.Nat Ecol Evol. 2024 May;8(5):1010-1020. doi: 10.1038/s41559-024-02367-y. Epub 2024 Mar 14. Nat Ecol Evol. 2024. PMID: 38486107 Free PMC article.
-
Emergence and maintenance of stable coexistence during a long-term multicellular evolution experiment.bioRxiv [Preprint]. 2023 Jan 20:2023.01.19.524803. doi: 10.1101/2023.01.19.524803. bioRxiv. 2023. Update in: Nat Ecol Evol. 2024 May;8(5):1010-1020. doi: 10.1038/s41559-024-02367-y. PMID: 36711513 Free PMC article. Updated. Preprint.
-
Why have aggregative multicellular organisms stayed simple?Curr Genet. 2021 Dec;67(6):871-876. doi: 10.1007/s00294-021-01193-0. Epub 2021 Jun 10. Curr Genet. 2021. PMID: 34114051 Review.
-
The evolution of multicellularity and cancer: views and paradigms.Biochem Soc Trans. 2020 Aug 28;48(4):1505-1518. doi: 10.1042/BST20190992. Biochem Soc Trans. 2020. PMID: 32677677 Review.
References
-
- Grosberg R. K., Strathmann R. R., The Evolution of Multicellularity: A Minor Major Transition? Annual Review of Ecology, Evolution, and Systematics 38, 621–654 (2007) DOI: 10.1146/annurev.ecolsys.36.102403.114735 - DOI
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
