Divergent structures of Mammalian and gammaherpesvirus uracil DNA glycosylases confer distinct DNA binding and substrate activity
- PMID: 37315375
- PMCID: PMC10441670
- DOI: 10.1016/j.dnarep.2023.103515
Divergent structures of Mammalian and gammaherpesvirus uracil DNA glycosylases confer distinct DNA binding and substrate activity
Abstract
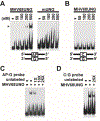
Uracil DNA glycosylase (UNG) removes mutagenic uracil base from DNA to initiate base excision repair (BER). The result is an abasic site (AP site) that is further processed by the high-fidelity BER pathway to complete repair and maintain genome integrity. The gammaherpesviruses (GHVs), human Kaposi sarcoma herpesvirus (KSHV), Epstein-Barr virus (EBV), and murine gammaherpesvirus 68 (MHV68) encode functional UNGs that have a role in viral genome replication. Mammalian and GHVs UNG share overall structure and sequence similarity except for a divergent amino-terminal domain and a leucine loop motif in the DNA binding domain that varies in sequence and length. To determine if divergent domains contribute to functional differences between GHV and mammalian UNGs, we analyzed their roles in DNA interaction and catalysis. By utilizing chimeric UNGs with swapped domains we found that the leucine loop in GHV, but not mammalian UNGs facilitates interaction with AP sites and that the amino-terminal domain modulates this interaction. We also found that the leucine loop structure contributes to differential UDGase activity on uracil in single- versus double-stranded DNA. Taken together we demonstrate that the GHV UNGs evolved divergent domains from their mammalian counterparts that contribute to differential biochemical properties from their mammalian counterparts.
Keywords: Abasic site; Gammaherpesvirus; Uracil DNA glycosylase.
Copyright © 2023 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


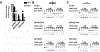


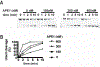
Similar articles
-
Uracil-DNA glycosylase of murine gammaherpesvirus 68 binds cognate viral replication factors independently of its catalytic residues.mSphere. 2023 Oct 24;8(5):e0027823. doi: 10.1128/msphere.00278-23. Epub 2023 Sep 25. mSphere. 2023. PMID: 37747202 Free PMC article.
-
New insights on the role of the gamma-herpesvirus uracil-DNA glycosylase leucine loop revealed by the structure of the Epstein-Barr virus enzyme in complex with an inhibitor protein.J Mol Biol. 2007 Feb 9;366(1):117-31. doi: 10.1016/j.jmb.2006.11.007. Epub 2006 Nov 7. J Mol Biol. 2007. PMID: 17157317
-
Characterization of the uracil-DNA glycosylase activity of Epstein-Barr virus BKRF3 and its role in lytic viral DNA replication.J Virol. 2007 Feb;81(3):1195-208. doi: 10.1128/JVI.01518-06. Epub 2006 Nov 15. J Virol. 2007. PMID: 17108049 Free PMC article.
-
Properties and functions of human uracil-DNA glycosylase from the UNG gene.Prog Nucleic Acid Res Mol Biol. 2001;68:365-86. doi: 10.1016/s0079-6603(01)68112-1. Prog Nucleic Acid Res Mol Biol. 2001. PMID: 11554311 Review.
-
Poxvirus uracil-DNA glycosylase-An unusual member of the family I uracil-DNA glycosylases.Protein Sci. 2016 Dec;25(12):2113-2131. doi: 10.1002/pro.3058. Epub 2016 Nov 2. Protein Sci. 2016. PMID: 27684934 Free PMC article. Review.
Cited by
-
Sequencing of Kaposi's Sarcoma Herpesvirus (KSHV) genomes from persons of diverse ethnicities and provenances with KSHV-associated diseases demonstrate multiple infections, novel polymorphisms, and low intra-host variance.PLoS Pathog. 2024 Jul 15;20(7):e1012338. doi: 10.1371/journal.ppat.1012338. eCollection 2024 Jul. PLoS Pathog. 2024. PMID: 39008527 Free PMC article.
-
Uracil-DNA glycosylase of murine gammaherpesvirus 68 binds cognate viral replication factors independently of its catalytic residues.mSphere. 2023 Oct 24;8(5):e0027823. doi: 10.1128/msphere.00278-23. Epub 2023 Sep 25. mSphere. 2023. PMID: 37747202 Free PMC article.
References
-
- Krokan HE, Saetrom P, Aas PA, Pettersen HS, Kavli B, Slupphaug G, Error-free versus mutagenic processing of genomic uracil--relevance to cancer, DNA Repair (Amst), 19 (2014) 38–47. - PubMed
-
- Safavi S, Larouche A, Zahn A, Patenaude AM, Domanska D, Dionne K, Rognes T, Dingler F, Kang SK, Liu Y, Johnson N, Hebert J, Verdun RE, Rada CA, Vega F, Nilsen H, Di Noia JM, The uracil-DNA glycosylase UNG protects the fitness of normal and cancer B cells expressing AID, NAR Cancer, 2 (2020) zcaa019. - PMC - PubMed
-
- Di Noia JM, Neuberger MS, Molecular mechanisms of antibody somatic hypermutation, Annu Rev Biochem, 76 (2007) 1–22. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

