Proteome architecture of human-induced pluripotent stem cell-derived three-dimensional organoids as a tool for early diagnosis of neuronal disorders
- PMID: 37313936
- PMCID: PMC10335645
- DOI: 10.4103/ijp.ijp_56_23
Proteome architecture of human-induced pluripotent stem cell-derived three-dimensional organoids as a tool for early diagnosis of neuronal disorders
Abstract
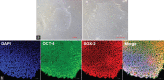
Background and objectives: Induced pluripotent stem cells (iPSCs) derived three-dimensional (3D) model for rare neurodegenerative disorders such as amyotrophic lateral sclerosis (ALS) is emerging as a novel alternative to human diseased tissue to explore the disease etiology and potential drug discovery. In the interest of the same, we have generated a TDP-43-mutated human iPSCs (hiPSCs) derived 3D organoid model of ALS disease. The high-resolution mass spectrometry (MS)-based proteomic approach is used to explore the differential mechanism under disease conditions and the suitability of a 3D model to study the disease.
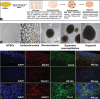
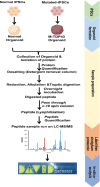
Materials and methods: The hiPSCs cell line was procured from a commercial source, grown, and characterized following standard protocols. The mutation in hiPSCs was accomplished using CRISPR/Cas-9 technology and predesigned gRNA. The two groups of organoids were produced by normal and mutated hiPSCs and subjected to the whole proteomic profiling by high-resolution MS in two biological replicates with three technical replicas of each.
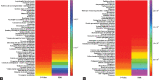
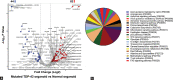
Results: The proteomic analysis of normal and mutated organoids revealed the proteins associated with pathways of neurodegenerative disorders, proteasomes, autophagy, and hypoxia-inducible factor-1 signaling. Differential proteomic analysis revealed that the mutation in TDP-43 gene caused proteomic deregulation, which impaired protein quality mechanisms. Furthermore, this impairment may contribute to the generation of stress conditions that may ultimately lead to the development of ALS pathology.
Conclusion: The developed 3D model represents the majority of candidate proteins and associated biological mechanisms altered in ALS disease. The study also offers novel protein targets that may uncloud the precise disease pathological mechanism and be considered for future diagnostic and therapeutic purposes for various neurodegenerative disorders.
Keywords: Amyotrophic lateral sclerosis; brain organoids; human-induced pluripotent stem cells; neurodegenerative disorders; proteome profiling.
Conflict of interest statement
None
Figures
Similar articles
-
Spinal cord extracts of amyotrophic lateral sclerosis spread TDP-43 pathology in cerebral organoids.PLoS Genet. 2023 Feb 6;19(2):e1010606. doi: 10.1371/journal.pgen.1010606. eCollection 2023 Feb. PLoS Genet. 2023. PMID: 36745687 Free PMC article.
-
3D brain Organoids derived from pluripotent stem cells: promising experimental models for brain development and neurodegenerative disorders.J Biomed Sci. 2017 Aug 20;24(1):59. doi: 10.1186/s12929-017-0362-8. J Biomed Sci. 2017. PMID: 28822354 Free PMC article. Review.
-
The proteomic architecture of schizophrenia iPSC-derived cerebral organoids reveals alterations in GWAS and neuronal development factors.Transl Psychiatry. 2021 Oct 19;11(1):541. doi: 10.1038/s41398-021-01664-5. Transl Psychiatry. 2021. PMID: 34667143 Free PMC article.
-
Label-Free LC-MS/MS Proteomic Analysis of Cerebrospinal Fluid Identifies Protein/Pathway Alterations and Candidate Biomarkers for Amyotrophic Lateral Sclerosis.J Proteome Res. 2015 Nov 6;14(11):4486-501. doi: 10.1021/acs.jproteome.5b00804. Epub 2015 Oct 8. J Proteome Res. 2015. PMID: 26401960 Free PMC article.
-
Reverse engineering human neurodegenerative disease using pluripotent stem cell technology.Brain Res. 2016 May 1;1638(Pt A):30-41. doi: 10.1016/j.brainres.2015.09.023. Epub 2015 Sep 28. Brain Res. 2016. PMID: 26423934 Free PMC article. Review.
Cited by
-
Morphogenetic Designs, and Disease Models in Central Nervous System Organoids.Int J Mol Sci. 2024 Jul 15;25(14):7750. doi: 10.3390/ijms25147750. Int J Mol Sci. 2024. PMID: 39062993 Free PMC article. Review.
-
A protein-miRNA biomic analysis approach to explore neuroprotective potential of nobiletin in human neural progenitor cells (hNPCs).Front Pharmacol. 2024 Jan 25;15:1343569. doi: 10.3389/fphar.2024.1343569. eCollection 2024. Front Pharmacol. 2024. PMID: 38348393 Free PMC article.
-
Proteomic-miRNA Biomics Profile Reveals 2D Cultures of Human iPSC-Derived Neural Progenitor Cells More Sensitive than 3D Spheroid System Against the Experimental Exposure to Arsenic.Mol Neurobiol. 2024 Aug;61(8):5754-5770. doi: 10.1007/s12035-024-03924-z. Epub 2024 Jan 16. Mol Neurobiol. 2024. PMID: 38228842
References
-
- Fujimori K, Ishikawa M, Otomo A, Atsuta N, Nakamura R, Akiyama T, et al. Modeling sporadic ALS in iPSC-derived motor neurons identifies a potential therapeutic agent. Nat Med. 2018;24:1579–89. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

