cGAS regulates the DNA damage response to maintain proliferative signaling in gastric cancer cells
- PMID: 37305397
- PMCID: PMC10208015
- DOI: 10.32604/or.2022.03529
cGAS regulates the DNA damage response to maintain proliferative signaling in gastric cancer cells
Abstract
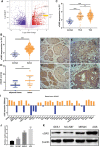
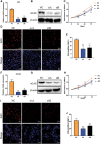
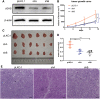
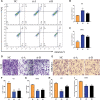
The activation of some oncogenes promote cancer cell proliferation and growth, facilitate cancer progression and metastasis by induce DNA replication stress, even genome instability. Activation of the cyclic GMP-AMP synthase (cGAS) mediates classical DNA sensing, is involved in genome instability, and is linked to various tumor development or therapy. However, the function of cGAS in gastric cancer remains elusive. In this study, the TCGA database and retrospective immunohistochemical analyses revealed substantially high cGAS expression in gastric cancer tissues and cell lines. By employing cGAS high-expression gastric cancer cell lines, including AGS and MKN45, ectopic silencing of cGAS caused a significant reduction in the proliferation of the cells, tumor growth, and mass in xenograft mice. Mechanistically, database analysis predicted a possible involvement of cGAS in the DNA damage response (DDR), further data through cells revealed protein interactions of the cGAS and MRE11-RAD50-NBN (MRN) complex, which activated cell cycle checkpoints, even increased genome instability in gastric cancer cells, thereby contributing to gastric cancer progression and sensitivity to treatment with DNA damaging agents. Furthermore, the upregulation of cGAS significantly exacerbated the prognosis of gastric cancer patients while improving radiotherapeutic outcomes. Therefore, we concluded that cGAS is involved in gastric cancer progression by fueling genome instability, implying that intervening in the cGAS pathway could be a practicable therapeutic approach for gastric cancer.
Keywords: Cell proliferation; DNA damage response; Gastric cancer; MRN complex; cGAS.
© 2021 Liu et al.
Conflict of interest statement
The authors have declared that they have no conflict of interest.
Figures



Similar articles
-
Identification and validation of the prognostic value of cyclic GMP-AMP synthase-stimulator of interferon (cGAS-STING) related genes in gastric cancer.Bioengineered. 2021 Dec;12(1):1238-1250. doi: 10.1080/21655979.2021.1911557. Bioengineered. 2021. PMID: 33843442 Free PMC article.
-
Targeting MUS81 promotes the anticancer effect of WEE1 inhibitor and immune checkpoint blocking combination therapy via activating cGAS/STING signaling in gastric cancer cells.J Exp Clin Cancer Res. 2021 Oct 8;40(1):315. doi: 10.1186/s13046-021-02120-4. J Exp Clin Cancer Res. 2021. PMID: 34625086 Free PMC article.
-
Expression of SASP, DNA Damage Response, and Cell Proliferation Factors in Early Gastric Neoplastic Lesions: Correlations and Clinical Significance.Pathol Oncol Res. 2022 Aug 19;28:1610401. doi: 10.3389/pore.2022.1610401. eCollection 2022. Pathol Oncol Res. 2022. PMID: 36061145 Free PMC article.
-
Regulation of cGAS-STING signalling in cancer: Approach for combination therapy.Biochim Biophys Acta Rev Cancer. 2023 May;1878(3):188896. doi: 10.1016/j.bbcan.2023.188896. Epub 2023 Apr 17. Biochim Biophys Acta Rev Cancer. 2023. PMID: 37088059 Review.
-
cGAS-STING signaling pathway in gastrointestinal inflammatory disease and cancers.FASEB J. 2022 Jan;36(1):e22029. doi: 10.1096/fj.202101199R. FASEB J. 2022. PMID: 34907606 Review.
Cited by
-
The role of cGAS in epithelial dysregulation in inflammatory bowel disease and gastrointestinal malignancies.Front Pharmacol. 2024 Jul 10;15:1409683. doi: 10.3389/fphar.2024.1409683. eCollection 2024. Front Pharmacol. 2024. PMID: 39050748 Free PMC article. Review.
-
CO2 reduction reaction on Sc-doped nanocages as catalysts.J Mol Model. 2023 Nov 20;29(12):381. doi: 10.1007/s00894-023-05776-1. J Mol Model. 2023. PMID: 37985487
References
-
- Siegel, R. L., Miller, K. D., Jemal, A. (2020). Cancer statistics, 2020. CA: A Cancer Journal for Clinicians , 70, 7–30. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
