Muscleblind-like proteins use modular domains to localize RNAs by riding kinesins and docking to membranes
- PMID: 37296096
- PMCID: PMC10256740
- DOI: 10.1038/s41467-023-38923-6
Muscleblind-like proteins use modular domains to localize RNAs by riding kinesins and docking to membranes
Abstract
RNA binding proteins (RBPs) act as critical facilitators of spatially regulated gene expression. Muscleblind-like (MBNL) proteins, implicated in myotonic dystrophy and cancer, localize RNAs to myoblast membranes and neurites through unknown mechanisms. We find that MBNL forms motile and anchored granules in neurons and myoblasts, and selectively associates with kinesins Kif1bα and Kif1c through its zinc finger (ZnF) domains. Other RBPs with similar ZnFs associate with these kinesins, implicating a motor-RBP specificity code. MBNL and kinesin perturbation leads to widespread mRNA mis-localization, including depletion of Nucleolin transcripts from neurites. Live cell imaging and fractionation reveal that the unstructured carboxy-terminal tail of MBNL1 allows for anchoring at membranes. An approach, termed RBP Module Recruitment and Imaging (RBP-MRI), reconstitutes kinesin- and membrane-recruitment functions using MBNL-MS2 coat protein fusions. Our findings decouple kinesin association, RNA binding, and membrane anchoring functions of MBNL while establishing general strategies for studying multi-functional, modular domains of RBPs.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
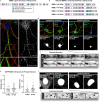

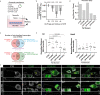
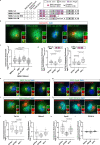



Similar articles
-
Muscleblind-Like 1 and Muscleblind-Like 3 Depletion Synergistically Enhances Myotonia by Altering Clc-1 RNA Translation.EBioMedicine. 2015 Jul 31;2(9):1034-47. doi: 10.1016/j.ebiom.2015.07.028. eCollection 2015 Sep. EBioMedicine. 2015. PMID: 26501102 Free PMC article.
-
Cognate RNA-Binding Modes by the Alternative-Splicing Regulator MBNL1 Inferred from Molecular Dynamics.Int J Mol Sci. 2022 Dec 18;23(24):16147. doi: 10.3390/ijms232416147. Int J Mol Sci. 2022. PMID: 36555788 Free PMC article.
-
Colocalization of muscleblind with RNA foci is separable from mis-regulation of alternative splicing in myotonic dystrophy.J Cell Sci. 2005 Jul 1;118(Pt 13):2923-33. doi: 10.1242/jcs.02404. Epub 2005 Jun 16. J Cell Sci. 2005. PMID: 15961406
-
The Muscleblind family of proteins: an emerging class of regulators of developmentally programmed alternative splicing.Differentiation. 2006 Mar;74(2-3):65-80. doi: 10.1111/j.1432-0436.2006.00060.x. Differentiation. 2006. PMID: 16533306 Review.
-
An Overview of Alternative Splicing Defects Implicated in Myotonic Dystrophy Type I.Genes (Basel). 2020 Sep 22;11(9):1109. doi: 10.3390/genes11091109. Genes (Basel). 2020. PMID: 32971903 Free PMC article. Review.
Cited by
-
MBNL splicing factors regulate the microtranscriptome of skeletal muscles.Nucleic Acids Res. 2024 Oct 28;52(19):12055-12073. doi: 10.1093/nar/gkae774. Nucleic Acids Res. 2024. PMID: 39258536 Free PMC article.
-
KIF1C facilitates retrograde transport of lysosomes through Hook3 and dynein.Commun Biol. 2024 Oct 11;7(1):1305. doi: 10.1038/s42003-024-07023-6. Commun Biol. 2024. PMID: 39394274 Free PMC article.
-
The kinesin-3 KIF1C undergoes liquid-liquid phase separation for accumulation of specific transcripts at the cell periphery.EMBO J. 2024 Aug;43(15):3192-3213. doi: 10.1038/s44318-024-00147-9. Epub 2024 Jun 19. EMBO J. 2024. PMID: 38898313 Free PMC article.
-
RNA in axons, dendrites, synapses and beyond.Front Mol Neurosci. 2024 Sep 18;17:1397378. doi: 10.3389/fnmol.2024.1397378. eCollection 2024. Front Mol Neurosci. 2024. PMID: 39359690 Free PMC article. Review.
-
hnRNP R promotes O-GlcNAcylation of eIF4G and facilitates axonal protein synthesis.Nat Commun. 2024 Aug 28;15(1):7430. doi: 10.1038/s41467-024-51678-y. Nat Commun. 2024. PMID: 39198412 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

