This is a preprint.
Pathogenic variants in Crx have distinct cis-regulatory effects on enhancers and silencers in photoreceptors
- PMID: 37292699
- PMCID: PMC10245955
- DOI: 10.1101/2023.05.27.542576
Pathogenic variants in Crx have distinct cis-regulatory effects on enhancers and silencers in photoreceptors
Update in
-
Pathogenic variants in CRX have distinct cis-regulatory effects on enhancers and silencers in photoreceptors.Genome Res. 2024 Mar 20;34(2):243-255. doi: 10.1101/gr.278133.123. Genome Res. 2024. PMID: 38355306 Free PMC article.
Abstract
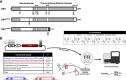
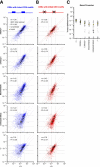
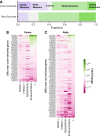
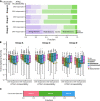
Dozens of variants in the photoreceptor-specific transcription factor (TF) CRX are linked with human blinding diseases that vary in their severity and age of onset. It is unclear how different variants in this single TF alter its function in ways that lead to a range of phenotypes. We examined the effects of human disease-causing variants on CRX cis-regulatory function by deploying massively parallel reporter assays (MPRAs) in live mouse retinas carrying knock-ins of two variants, one in the DNA binding domain (p.R90W) and the other in the transcriptional effector domain (p.E168d2). The degree of reporter gene dysregulation caused by the variants corresponds with their phenotypic severity. The two variants affect similar sets of enhancers, while p.E168d2 has stronger effects on silencers. Cis-regulatory elements (CREs) near cone photoreceptor genes are enriched for silencers that are de-repressed in the presence of p.E168d2. Chromatin environments of CRX-bound loci were partially predictive of episomal MPRA activity, and silencers were notably enriched among distal elements whose accessibility increases later in retinal development. We identified a set of potentially pleiotropic regulatory elements that convert from silencers to enhancers in retinas that lack a functional CRX effector domain. Our findings show that phenotypically distinct variants in different domains of CRX have partially overlapping effects on its cis-regulatory function, leading to misregulation of similar sets of enhancers, while having a qualitatively different impact on silencers.
Conflict of interest statement
Competing Interest Statement B.A.C is on the scientific advisory board of Patch Biosciences. The authors declare no other competing interests.
Figures


Similar articles
-
Pathogenic variants in CRX have distinct cis-regulatory effects on enhancers and silencers in photoreceptors.Genome Res. 2024 Mar 20;34(2):243-255. doi: 10.1101/gr.278133.123. Genome Res. 2024. PMID: 38355306 Free PMC article.
-
Information content differentiates enhancers from silencers in mouse photoreceptors.Elife. 2021 Sep 6;10:e67403. doi: 10.7554/eLife.67403. Elife. 2021. PMID: 34486522 Free PMC article.
-
Mechanistically distinct mouse models for CRX-associated retinopathy.PLoS Genet. 2014 Feb 6;10(2):e1004111. doi: 10.1371/journal.pgen.1004111. eCollection 2014 Feb. PLoS Genet. 2014. PMID: 24516401 Free PMC article.
-
High-Throughput Analysis of Retinal Cis-Regulatory Networks by Massively Parallel Reporter Assays.Adv Exp Med Biol. 2019;1185:359-364. doi: 10.1007/978-3-030-27378-1_59. Adv Exp Med Biol. 2019. PMID: 31884638 Free PMC article. Review.
-
Transcriptional Silencers: Driving Gene Expression with the Brakes On.Trends Genet. 2021 Jun;37(6):514-527. doi: 10.1016/j.tig.2021.02.002. Epub 2021 Mar 9. Trends Genet. 2021. PMID: 33712326 Free PMC article. Review.
References
-
- Bailey TL, Grant CE. 2021. SEA: Simple Enrichment Analysis of motifs. bioRxiv.
-
- Berger W, Kloeckener-Gruissem B, Neidhardt J. 2010. The molecular basis of human retinal and vitreoretinal diseases. Progress in Retinal and Eye Research 29: 335–375. - PubMed
-
- Chau K-Y, Chen S, Zack DJ, Ono SJ. 2000. Functional Domains of the Cone-Rod Homeobox (CRX) Transcription Factor. Journal of Biological Chemistry 275: 37264–37270. - PubMed
-
- Chen S, Wang Q-L, Nie Z, Sun H, Lennon G, Copeland NG, Gilbert DJ, Jenkins NA, Zack DJ. 1997. Crx, a Novel Otx-like Paired-Homeodomain Protein, Binds to and Transactivates Photoreceptor Cell-Specific Genes. Neuron 19: 1017–1030. - PubMed
-
- Chen S, Wang Q-L, Xu S, Liu I, Li LY, Wang Y, Zack DJ. 2002. Functional analysis of conerod homeobox (CRX) mutations associated with retinal dystrophy. Human Molecular Genetics 11: 873–884. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
