FAM family gene prediction model reveals heterogeneity, stemness and immune microenvironment of UCEC
- PMID: 37275958
- PMCID: PMC10235772
- DOI: 10.3389/fmolb.2023.1200335
FAM family gene prediction model reveals heterogeneity, stemness and immune microenvironment of UCEC
Abstract
Background: Endometrial cancer (UCEC) is a highly heterogeneous gynecologic malignancy that exhibits variable prognostic outcomes and responses to immunotherapy. The Familial sequence similarity (FAM) gene family is known to contribute to the pathogenesis of various malignancies, but the extent of their involvement in UCEC has not been systematically studied. This investigation aimed to develop a robust risk profile based on FAM family genes (FFGs) to predict the prognosis and suitability for immunotherapy in UCEC patients. Methods: Using the TCGA-UCEC cohort from The Cancer Genome Atlas (TCGA) database, we obtained expression profiles of FFGs from 552 UCEC and 35 normal samples, and analyzed the expression patterns and prognostic relevance of 363 FAM family genes. The UCEC samples were randomly divided into training and test sets (1:1), and univariate Cox regression analysis and Lasso Cox regression analysis were conducted to identify the differentially expressed genes (FAM13C, FAM110B, and FAM72A) that were significantly associated with prognosis. A prognostic risk scoring system was constructed based on these three gene characteristics using multivariate Cox proportional risk regression. The clinical potential and immune status of FFGs were analyzed using CiberSort, SSGSEA, and tumor immune dysfunction and rejection (TIDE) algorithms. qRT-PCR and IHC for detecting the expression levels of 3-FFGs. Results: Three FFGs, namely, FAM13C, FAM110B, and FAM72A, were identified as strongly associated with the prognosis of UCEC and effective predictors of UCEC prognosis. Multivariate analysis demonstrated that the developed model was an independent predictor of UCEC, and that patients in the low-risk group had better overall survival than those in the high-risk group. The nomogram constructed from clinical characteristics and risk scores exhibited good prognostic power. Patients in the low-risk group exhibited a higher tumor mutational load (TMB) and were more likely to benefit from immunotherapy. Conclusion: This study successfully developed and validated novel biomarkers based on FFGs for predicting the prognosis and immune status of UCEC patients. The identified FFGs can accurately assess the prognosis of UCEC patients and facilitate the identification of specific subgroups of patients who may benefit from personalized treatment with immunotherapy and chemotherapy.
Keywords: FAM family genes; UCEC; cancer treatment; chemotherapy; stemness; tumor heterogeneity; tumor microenvironment.
Copyright © 2023 Chi, Gao, Xia, Yu, Yin, Pan, Peng, Mao, Teichmann, Zhang, Tran, Jiang, Liu, Yang and Wang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

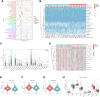


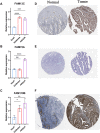
Similar articles
-
Integrated bioinformatics analysis and experimental validation reveals fatty acid metabolism-related prognostic signature and immune responses for uterine corpus endometrial carcinoma.Front Oncol. 2022 Nov 9;12:1030246. doi: 10.3389/fonc.2022.1030246. eCollection 2022. Front Oncol. 2022. PMID: 36439473 Free PMC article.
-
Development and Clinical Validation of Novel 8-Gene Prognostic Signature Associated With the Proportion of Regulatory T Cells by Weighted Gene Co-Expression Network Analysis in Uterine Corpus Endometrial Carcinoma.Front Immunol. 2021 Dec 14;12:788431. doi: 10.3389/fimmu.2021.788431. eCollection 2021. Front Immunol. 2021. PMID: 34970268 Free PMC article.
-
Identification of methylation-driven genes prognosis signature and immune microenvironment in uterus corpus endometrial cancer.Cancer Cell Int. 2021 Jul 10;21(1):365. doi: 10.1186/s12935-021-02038-z. Cancer Cell Int. 2021. PMID: 34246261 Free PMC article.
-
Prognostic signature construction and immunotherapy response analysis for Uterine Corpus Endometrial Carcinoma based on cuproptosis-related lncRNAs.Comput Biol Med. 2023 Jun;159:106905. doi: 10.1016/j.compbiomed.2023.106905. Epub 2023 Apr 11. Comput Biol Med. 2023. PMID: 37060773
-
Construction of N-7 methylguanine-related mRNA prognostic model in uterine corpus endometrial carcinoma based on multi-omics data and immune-related analysis.Sci Rep. 2022 Nov 5;12(1):18813. doi: 10.1038/s41598-022-22879-6. Sci Rep. 2022. PMID: 36335189 Free PMC article.
Cited by
-
Pan-cancer analysis and single-cell analysis reveals FAM110B as a potential target for survival and immunotherapy.Front Mol Biosci. 2024 Aug 7;11:1424104. doi: 10.3389/fmolb.2024.1424104. eCollection 2024. Front Mol Biosci. 2024. PMID: 39170745 Free PMC article.
-
Unveiling efferocytosis-related signatures through the integration of single-cell analysis and machine learning: a predictive framework for prognosis and immunotherapy response in hepatocellular carcinoma.Front Immunol. 2023 Jul 27;14:1237350. doi: 10.3389/fimmu.2023.1237350. eCollection 2023. Front Immunol. 2023. PMID: 37575252 Free PMC article.
-
Identification of M5c regulator-medicated methylation modification patterns for prognosis and immune microenvironment in glioma.Aging (Albany NY). 2023 Nov 6;15(21):12275-12295. doi: 10.18632/aging.205179. Epub 2023 Nov 6. Aging (Albany NY). 2023. PMID: 37934565 Free PMC article.
-
Deciphering the role of tryptophan metabolism-associated genes ECHS1 and ALDH2 in gastric cancer: implications for tumor immunity and personalized therapy.Front Immunol. 2024 Sep 12;15:1460308. doi: 10.3389/fimmu.2024.1460308. eCollection 2024. Front Immunol. 2024. PMID: 39328412 Free PMC article.
-
Integrating single-cell and multi-omic approaches reveals Euphorbiae Humifusae Herba-dependent mitochondrial dysfunction in non-small-cell lung cancer.J Cell Mol Med. 2024 May;28(10):e18317. doi: 10.1111/jcmm.18317. J Cell Mol Med. 2024. PMID: 38801409 Free PMC article.
References
-
- Ayesha M., Majid A., Zhao D., Greenaway F. T., Yan N., Liu Q., et al. (2022). MiR-4521 plays a tumor repressive role in growth and metastasis of hepatocarcinoma cells by suppressing phosphorylation of FAK/AKT pathway via targeting FAM129A. J. Adv. Res. 36, 147–161. 10.1016/j.jare.2021.05.003 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

