Halophytes as new model plant species for salt tolerance strategies
- PMID: 37251767
- PMCID: PMC10211249
- DOI: 10.3389/fpls.2023.1137211
Halophytes as new model plant species for salt tolerance strategies
Abstract
Soil salinity is becoming a growing issue nowadays, severely affecting the world's most productive agricultural landscapes. With intersecting and competitive challenges of shrinking agricultural lands and increasing demand for food, there is an emerging need to build resilience for adaptation to anticipated climate change and land degradation. This necessitates the deep decoding of a gene pool of crop plant wild relatives which can be accomplished through salt-tolerant species, such as halophytes, in order to reveal the underlying regulatory mechanisms. Halophytes are generally defined as plants able to survive and complete their life cycle in highly saline environments of at least 200-500 mM of salt solution. The primary criterion for identifying salt-tolerant grasses (STGs) includes the presence of salt glands on the leaf surface and the Na+ exclusion mechanism since the interaction and replacement of Na+ and K+ greatly determines the survivability of STGs in saline environments. During the last decades or so, various salt-tolerant grasses/halophytes have been explored for the mining of salt-tolerant genes and testing their efficacy to improve the limit of salt tolerance in crop plants. Still, the utility of halophytes is limited due to the non-availability of any model halophytic plant system as well as the lack of complete genomic information. To date, although Arabidopsis (Arabidopsis thaliana) and salt cress (Thellungiella halophila) are being used as model plants in most salt tolerance studies, these plants are short-lived and can tolerate salinity for a shorter duration only. Thus, identifying the unique genes for salt tolerance pathways in halophytes and their introgression in a related cereal genome for better tolerance to salinity is the need of the hour. Modern technologies including RNA sequencing and genome-wide mapping along with advanced bioinformatics programs have advanced the decoding of the whole genetic information of plants and the development of probable algorithms to correlate stress tolerance limit and yield potential. Hence, this article has been compiled to explore the naturally occurring halophytes as potential model plant species for abiotic stress tolerance and to further breed crop plants to enhance salt tolerance through genomic and molecular tools.
Keywords: DEGs (differentially expressed genes); gene transformation; halophytes; osmoregulation; salinity; salt tolerance; transcriptomics.
Copyright © 2023 Mann, Lata, Kumar, Kumar, Kumar and Sheoran.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
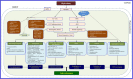

Similar articles
-
Salt tolerance mechanisms in Salt Tolerant Grasses (STGs) and their prospects in cereal crop improvement.Bot Stud. 2014 Dec;55(1):31. doi: 10.1186/1999-3110-55-31. Epub 2014 Mar 14. Bot Stud. 2014. PMID: 28510965 Free PMC article. Review.
-
Comparative 2D-DIGE analysis of salinity responsive microsomal proteins from leaves of salt-sensitive Arabidopsis thaliana and salt-tolerant Thellungiella salsuginea.J Proteomics. 2014 Dec 5;111:113-27. doi: 10.1016/j.jprot.2014.05.018. Epub 2014 Jun 2. J Proteomics. 2014. PMID: 24892798
-
Adaptive Mechanisms of Halophytes and Their Potential in Improving Salinity Tolerance in Plants.Int J Mol Sci. 2021 Oct 3;22(19):10733. doi: 10.3390/ijms221910733. Int J Mol Sci. 2021. PMID: 34639074 Free PMC article. Review.
-
Balancing salinity stress responses in halophytes and non-halophytes: a comparison between Thellungiella and Arabidopsis thaliana.Funct Plant Biol. 2013 Aug;40(9):819-831. doi: 10.1071/FP12299. Funct Plant Biol. 2013. PMID: 32481153
-
A comparative study of salt tolerance parameters in 11 wild relatives of Arabidopsis thaliana.J Exp Bot. 2010 Aug;61(13):3787-98. doi: 10.1093/jxb/erq188. Epub 2010 Jul 1. J Exp Bot. 2010. PMID: 20595237 Free PMC article.
Cited by
-
Understanding of Plant Salt Tolerance Mechanisms and Application to Molecular Breeding.Int J Mol Sci. 2024 Oct 11;25(20):10940. doi: 10.3390/ijms252010940. Int J Mol Sci. 2024. PMID: 39456729 Free PMC article. Review.
-
Cakile maritima: A Halophyte Model to Study Salt Tolerance Mechanisms and Potential Useful Crop for Sustainable Saline Agriculture in the Context of Climate Change.Plants (Basel). 2024 Oct 15;13(20):2880. doi: 10.3390/plants13202880. Plants (Basel). 2024. PMID: 39458826 Free PMC article. Review.
-
La (NO3)3 substantially fortified Glycyrrhiza uralensis's resilience against salt stress by interconnected pathways.BMC Plant Biol. 2024 Oct 5;24(1):926. doi: 10.1186/s12870-024-05644-x. BMC Plant Biol. 2024. PMID: 39367329 Free PMC article.
-
Role of leaf micro-structural modifications in modulation of growth and photosynthetic performance of aquatic halophyte Fimbristylis complanata (Retz.) under temporal salinity regimes.Sci Rep. 2024 Nov 2;14(1):26442. doi: 10.1038/s41598-024-77589-y. Sci Rep. 2024. PMID: 39488568 Free PMC article.
-
Unveiling the influence of salinity on bacterial microbiome assembly of halophytes and crops.Environ Microbiome. 2024 Jul 18;19(1):49. doi: 10.1186/s40793-024-00592-3. Environ Microbiome. 2024. PMID: 39026296 Free PMC article.
References
-
- Aliakbari M., Razi H., Alemzadeh A., Tavakol E. (2020). RNA-Seq transcriptome profiling of the halophyte Salicornia persica in response to salinity. J. Plant Growth Regul. 40, 707–721. doi: 10.1007/s00344-020-10134-z - DOI
-
- Altuntas C., Terzi R. (2021). Concomitant accumulations of ions, osmoprotectants and antioxidant system-related substances provide salt tolerance capability to succulent extreme halophyte Scorzonera hieraciifolia . Turk. J. Bot. 45, 340–352. doi: 10.3906/bot-2102-7 - DOI
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

