Lipid acyl chain protrusion induced by the influenza virus hemagglutinin fusion peptide detected by NMR paramagnetic relaxation enhancement
- PMID: 37247572
- PMCID: PMC10330521
- DOI: 10.1016/j.bpc.2023.107028
Lipid acyl chain protrusion induced by the influenza virus hemagglutinin fusion peptide detected by NMR paramagnetic relaxation enhancement
Abstract
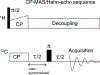
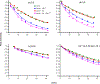
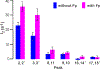
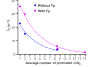
The glycoprotein spikes of membrane-enveloped viruses include a subunit that catalyzes fusion (joining) of the viral and target cell membranes. For influenza virus, this is subunit 2 of hemagglutinin which has a ∼ 20-residue N-terminal fusion peptide (Fp) region that binds target membrane. An outstanding question is whether there are associated membrane changes important for fusion. Several computational studies have found increased "protrusion" of lipid acyl chains near Fp, i.e. one or more chain carbons are closer to the aqueous region than the headgroup phosphorus. Protrusion may accelerate initial joining of outer leaflets of the two membranes into a stalk intermediate. In this study, higher protrusion probability in membrane with vs. without Fp is convincingly detected by larger Mn2+-associated increases in chain 13C NMR transverse relaxation rates (Γ2's). Data analysis provides a ratio Γ2,neighbor/Γ2,distant for lipids neighboring vs. more distant from the Fp. The calculated ratio depends on the number of Fp-neighboring lipids and the experimentally-derived range of 4 to 24 matches the range of increased protrusion probabilities from different simulations. For samples either with or without Fp, the Γ2 values are well-fitted by an exponential decay as the 13C site moves closer to the chain terminus. The decays correlate with free-energy of protrusion proportional to the number of protruded -CH2 groups, with free energy per -CH2 of ∼0.25 kBT. The NMR data support one major fusion role of the Fp to be much greater protrusion of lipid chains, with highest protrusion probability for chain regions closest to the headgroups.
Keywords: Fusion peptide; Hemagglutinin; Influenza; NMR; PRE; Protrusion.
Copyright © 2023 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
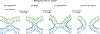
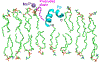
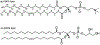


Similar articles
-
Rapid 2H NMR Transverse Relaxation of Perdeuterated Lipid Acyl Chains of Membrane with Bound Viral Fusion Peptide Supports Large-Amplitude Motions of These Chains That Can Catalyze Membrane Fusion.Biochemistry. 2021 Sep 7;60(35):2637-2651. doi: 10.1021/acs.biochem.1c00316. Epub 2021 Aug 26. Biochemistry. 2021. PMID: 34436856 Free PMC article.
-
2H nuclear magnetic resonance spectroscopy supports larger amplitude fast motion and interference with lipid chain ordering for membrane that contains β sheet human immunodeficiency virus gp41 fusion peptide or helical hairpin influenza virus hemagglutinin fusion peptide at fusogenic pH.Biochim Biophys Acta Biomembr. 2020 Oct 1;1862(10):183404. doi: 10.1016/j.bbamem.2020.183404. Epub 2020 Jun 23. Biochim Biophys Acta Biomembr. 2020. PMID: 32585207 Free PMC article.
-
The influenza fusion peptide promotes lipid polar head intrusion through hydrogen bonding with phosphates and N-terminal membrane insertion depth.Proteins. 2014 Sep;82(9):2118-27. doi: 10.1002/prot.24568. Epub 2014 Apr 16. Proteins. 2014. PMID: 24668589
-
Membrane fusion mechanisms: the influenza hemagglutinin paradigm and its implications for intracellular fusion.Traffic. 2000 Aug;1(8):598-604. doi: 10.1034/j.1600-0854.2000.010803.x. Traffic. 2000. PMID: 11208147 Review.
-
What studies of fusion peptides tell us about viral envelope glycoprotein-mediated membrane fusion (review).Mol Membr Biol. 1997 Jul-Sep;14(3):97-112. doi: 10.3109/09687689709048170. Mol Membr Biol. 1997. PMID: 9394290 Review.
Cited by
-
Visualizing intermediate stages of viral membrane fusion by cryo-electron tomography.Trends Biochem Sci. 2024 Oct;49(10):916-931. doi: 10.1016/j.tibs.2024.06.012. Epub 2024 Jul 24. Trends Biochem Sci. 2024. PMID: 39054240 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

