Fluorescence Microscopy in Adeno-Associated Virus Research
- PMID: 37243260
- PMCID: PMC10222864
- DOI: 10.3390/v15051174
Fluorescence Microscopy in Adeno-Associated Virus Research
Abstract
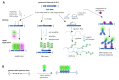
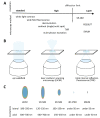
Research on adeno-associated virus (AAV) and its recombinant vectors as well as on fluorescence microscopy imaging is rapidly progressing driven by clinical applications and new technologies, respectively. The topics converge, since high and super-resolution microscopes facilitate the study of spatial and temporal aspects of cellular virus biology. Labeling methods also evolve and diversify. We review these interdisciplinary developments and provide information on the technologies used and the biological knowledge gained. The emphasis lies on the visualization of AAV proteins by chemical fluorophores, protein fusions and antibodies as well as on methods for the detection of adeno-associated viral DNA. We add a short overview of fluorescent microscope techniques and their advantages and challenges in detecting AAV.
Keywords: AAV; AAV labeling; DNA labeling; adeno-associated virus; laser scanning confocal microscopy (LSCM); microscopy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Adeno-associated virus: from defective virus to effective vector.Virol J. 2005 May 6;2:43. doi: 10.1186/1743-422X-2-43. Virol J. 2005. PMID: 15877812 Free PMC article. Review.
-
Adeno-associated virus biology.Methods Mol Biol. 2011;807:1-23. doi: 10.1007/978-1-61779-370-7_1. Methods Mol Biol. 2011. PMID: 22034024
-
Green fluorescent protein-tagged adeno-associated virus particles allow the study of cytosolic and nuclear trafficking.J Virol. 2005 Sep;79(18):11776-87. doi: 10.1128/JVI.79.18.11776-11787.2005. J Virol. 2005. PMID: 16140755 Free PMC article.
-
Herpes simplex virus type 1/adeno-associated virus hybrid vectors mediate site-specific integration at the adeno-associated virus preintegration site, AAVS1, on human chromosome 19.J Virol. 2002 Jul;76(14):7163-73. doi: 10.1128/jvi.76.14.7163-7173.2002. J Virol. 2002. PMID: 12072516 Free PMC article.
-
The state of the art of adeno-associated virus-based vectors in gene therapy.Virol J. 2007 Oct 16;4:99. doi: 10.1186/1743-422X-4-99. Virol J. 2007. PMID: 17939872 Free PMC article. Review.
Cited by
-
Exploring HIV-1 Maturation: A New Frontier in Antiviral Development.Viruses. 2024 Sep 6;16(9):1423. doi: 10.3390/v16091423. Viruses. 2024. PMID: 39339899 Free PMC article. Review.
-
Open-source and low-cost miniature microscope for on-site fluorescence detection.HardwareX. 2024 Jun 15;19:e00545. doi: 10.1016/j.ohx.2024.e00545. eCollection 2024 Sep. HardwareX. 2024. PMID: 39006472 Free PMC article.
-
A novel approach to quantitate biodistribution and transduction of adeno-associated virus gene therapy using radiolabeled AAV vectors in mice.Mol Ther Methods Clin Dev. 2024 Aug 19;32(3):101326. doi: 10.1016/j.omtm.2024.101326. eCollection 2024 Sep 12. Mol Ther Methods Clin Dev. 2024. PMID: 39286334 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

