GPR19 Coordinates Multiple Molecular Aspects of Stress Responses Associated with the Aging Process
- PMID: 37239845
- PMCID: PMC10218176
- DOI: 10.3390/ijms24108499
GPR19 Coordinates Multiple Molecular Aspects of Stress Responses Associated with the Aging Process
Abstract
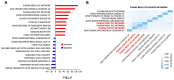
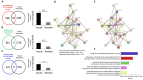
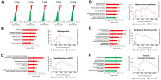
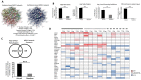
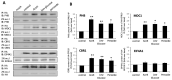
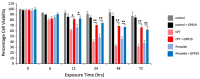
G protein-coupled receptors (GPCRs) play a significant role in controlling biological paradigms such as aging and aging-related disease. We have previously identified receptor signaling systems that are specifically associated with controlling molecular pathologies associated with the aging process. Here, we have identified a pseudo-orphan GPCR, G protein-coupled receptor 19 (GPR19), that is sensitive to many molecular aspects of the aging process. Through an in-depth molecular investigation process that involved proteomic, molecular biological, and advanced informatic experimentation, this study found that the functionality of GPR19 is specifically linked to sensory, protective, and remedial signaling systems associated with aging-related pathology. This study suggests that the activity of this receptor may play a role in mitigating the effects of aging-related pathology by promoting protective and remedial signaling systems. GPR19 expression variation demonstrates variability in the molecular activity in this larger process. At low expression levels in HEK293 cells, GPR19 expression regulates signaling paradigms linked with stress responses and metabolic responses to these. At higher expression levels, GPR19 expression co-regulates systems involved in sensing and repairing DNA damage, while at the highest levels of GPR19 expression, a functional link to processes of cellular senescence is seen. In this manner, GPR19 may function as a coordinator of aging-associated metabolic dysfunction, stress response, DNA integrity management, and eventual senescence.
Keywords: DNA; GPR19; adiposity; aging; damage; longevity; metabolism; mitochondria; receptor; stress.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
Intersection of the Orphan G Protein-Coupled Receptor, GPR19, with the Aging Process.Int J Mol Sci. 2022 Nov 6;23(21):13598. doi: 10.3390/ijms232113598. Int J Mol Sci. 2022. PMID: 36362387 Free PMC article. Review.
-
G protein-coupled receptor GPR19 regulates E-cadherin expression and invasion of breast cancer cells.Biochim Biophys Acta Mol Cell Res. 2017 Jul;1864(7):1318-1327. doi: 10.1016/j.bbamcr.2017.05.001. Epub 2017 May 2. Biochim Biophys Acta Mol Cell Res. 2017. PMID: 28476646
-
Expression of G protein-coupled receptor 19 in human lung cancer cells is triggered by entry into S-phase and supports G(2)-M cell-cycle progression.Mol Cancer Res. 2012 Oct;10(10):1343-58. doi: 10.1158/1541-7786.MCR-12-0139. Epub 2012 Aug 21. Mol Cancer Res. 2012. PMID: 22912338
-
The orphan G-protein-coupled receptor GPR19 is expressed predominantly in neuronal cells during mouse embryogenesis.Cell Tissue Res. 2004 Nov;318(2):459-63. doi: 10.1007/s00441-004-0948-9. Epub 2004 Sep 28. Cell Tissue Res. 2004. PMID: 15452705
-
G Protein-Coupled Receptor Systems as Crucial Regulators of DNA Damage Response Processes.Int J Mol Sci. 2018 Sep 26;19(10):2919. doi: 10.3390/ijms19102919. Int J Mol Sci. 2018. PMID: 30261591 Free PMC article. Review.
References
-
- Maudsley S., Martin B., Janssens J., Etienne H., Jushaj A., van Gastel J., Willemsen A., Chen H., Gesty-Palmer D., Luttrell L.M. Informatic deconvolution of biased GPCR signaling mechanisms from in vivo pharmacological experimentation. Methods. 2016;92:51–63. doi: 10.1016/j.ymeth.2015.05.013. - DOI - PMC - PubMed
-
- Maudsley S., Leysen H., van Gastel J., Martin B. Systems Pharmacology: Enabling Multidimensional Therapeutics. Elsevier; Amsterdam, The Netherlands: 2021. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

