Structural Basis of PE_PGRS Polymorphism, a Tool for Functional Modulation
- PMID: 37238682
- PMCID: PMC10216338
- DOI: 10.3390/biom13050812
Structural Basis of PE_PGRS Polymorphism, a Tool for Functional Modulation
Abstract
Background: The mycobacterial PE_PGRS protein family is present only in pathogenic strains of the genus mycobacterium, such as Mtb and members of the MTB complex, suggesting a likely important role of this family in pathogenesis. Their PGRS domains are highly polymorphic and have been suggested to cause antigenic variations and facilitate pathogen survival. The availability of AlphaFold2.0 offered us a unique opportunity to better understand structural and functional properties of these domains and a role of polymorphism in Mtb evolution and dissemination.
Methods: We made extensive use of AlphaFold2.0 computations and coupled them with sequence distribution phylogenetic and frequency analyses, and antigenic predictions.
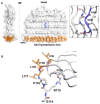
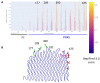
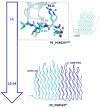
Results: Modeling of several polymorphic forms of PE_PGRS33, the prototype of the PE_PGRS family and sequence analyses allowed us to predict the structural impact of mutations/deletions/insertions present in the most frequent variants. These analyses well correlate with the observed frequency and with the phenotypic features of the described variants.
Conclusions: Here, we provide a thorough description of structural impacts of the observed polymorphism of PE_PGRS33 protein and we correlate predicted structures to the known fitness of strains containing specific variants. Finally, we also identify protein variants associated with bacterial evolution, showing sophisticated modifications likely endowed with a gain-of-function role during bacterial evolution.
Keywords: PE_PGRS; polymorphism; protein structure; tuberculosis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





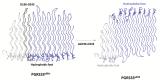

Similar articles
-
Impact of pe_pgrs33 Gene Polymorphisms on Mycobacterium tuberculosis Infection and Pathogenesis.Front Cell Infect Microbiol. 2017 Apr 21;7:137. doi: 10.3389/fcimb.2017.00137. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28484686 Free PMC article.
-
Execution of macrophage apoptosis by PE_PGRS33 of Mycobacterium tuberculosis is mediated by Toll-like receptor 2-dependent release of tumor necrosis factor-alpha.J Biol Chem. 2007 Jan 12;282(2):1039-50. doi: 10.1074/jbc.M604379200. Epub 2006 Nov 9. J Biol Chem. 2007. PMID: 17095513
-
Mycobacterium tuberculosis PE_PGRS16 and PE_PGRS26 genetic polymorphism among clinical isolates.Tuberculosis (Edinb). 2008 Jul;88(4):283-94. doi: 10.1016/j.tube.2008.01.001. Epub 2008 Mar 4. Tuberculosis (Edinb). 2008. PMID: 18313360 Free PMC article.
-
PE_PGRS proteins of Mycobacterium tuberculosis: A specialized molecular task force at the forefront of host-pathogen interaction.Virulence. 2020 Dec;11(1):898-915. doi: 10.1080/21505594.2020.1785815. Virulence. 2020. PMID: 32713249 Free PMC article. Review.
-
An overview to understand the role of PE_PGRS family proteins in Mycobacterium tuberculosis H37 Rv and their potential as new drug targets.Biotechnol Appl Biochem. 2015 Mar-Apr;62(2):145-53. doi: 10.1002/bab.1266. Epub 2014 Nov 11. Biotechnol Appl Biochem. 2015. PMID: 24975480 Review.
References
-
- Gey van Pittius N.C., Sampson S.L., Lee H., Kim Y., van Helden P.D., Warren R.M. Evolution and Expansion of the Mycobacterium Tuberculosis PE and PPE Multigene Families and Their Association with the Duplication of the ESAT-6 (Esx) Gene Cluster Regions. BMC Evol. Biol. 2006;6:95. doi: 10.1186/1471-2148-6-95. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

