The Ubiquitin-like Proteins of Saccharomyces cerevisiae
- PMID: 37238603
- PMCID: PMC10216172
- DOI: 10.3390/biom13050734
The Ubiquitin-like Proteins of Saccharomyces cerevisiae
Abstract
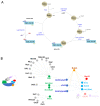
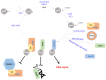
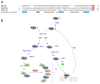
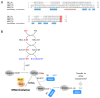
In this review, we present a comprehensive list of the ubiquitin-like modifiers (Ubls) of Saccharomyces cerevisiae, a common model organism used to study fundamental cellular processes that are conserved in complex multicellular organisms, such as humans. Ubls are a family of proteins that share structural relationships with ubiquitin, and which modify target proteins and lipids. These modifiers are processed, activated and conjugated to substrates by cognate enzymatic cascades. The attachment of substrates to Ubls alters the various properties of these substrates, such as function, interaction with the environment or turnover, and accordingly regulate key cellular processes, including DNA damage, cell cycle progression, metabolism, stress response, cellular differentiation, and protein homeostasis. Thus, it is not surprising that Ubls serve as tools to study the underlying mechanism involved in cellular health. We summarize current knowledge on the activity and mechanism of action of the S. cerevisiae Rub1, Smt3, Atg8, Atg12, Urm1 and Hub1 modifiers, all of which are highly conserved in organisms from yeast to humans.
Keywords: Atg12; Atg8; Hub1; NEDDylation; Rub1; S. cerevisiae; SUMOylation; Smt3; Ubls; Urm1; proteostasis; ubiquitin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Ubiquitin-like modifications in the DNA damage response.Mutat Res. 2017 Oct;803-805:56-75. doi: 10.1016/j.mrfmmm.2017.07.001. Epub 2017 Jul 11. Mutat Res. 2017. PMID: 28734548 Review.
-
Urm1: A Non-Canonical UBL.Biomolecules. 2021 Jan 22;11(2):139. doi: 10.3390/biom11020139. Biomolecules. 2021. PMID: 33499055 Free PMC article. Review.
-
The ubiquitin-like protein HUB1 forms SDS-resistant complexes with cellular proteins in the absence of ATP.EMBO Rep. 2003 Dec;4(12):1169-74. doi: 10.1038/sj.embor.7400025. Epub 2003 Nov 7. EMBO Rep. 2003. PMID: 14608371 Free PMC article.
-
The Proteasome Lid Triggers COP9 Signalosome Activity during the Transition of Saccharomyces cerevisiae Cells into Quiescence.Biomolecules. 2019 Sep 4;9(9):449. doi: 10.3390/biom9090449. Biomolecules. 2019. PMID: 31487956 Free PMC article.
-
Recognition and cleavage of related to ubiquitin 1 (Rub1) and Rub1-ubiquitin chains by components of the ubiquitin-proteasome system.Mol Cell Proteomics. 2012 Dec;11(12):1595-611. doi: 10.1074/mcp.M112.022467. Epub 2012 Oct 26. Mol Cell Proteomics. 2012. PMID: 23105008 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

