Overview of distinct N6-Methyladenosine profiles of messenger RNA in osteoarthritis
- PMID: 37229206
- PMCID: PMC10203613
- DOI: 10.3389/fgene.2023.1168365
Overview of distinct N6-Methyladenosine profiles of messenger RNA in osteoarthritis
Abstract
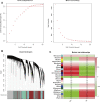
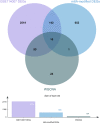
Although N6-methyladenosine (m6A) modification is closely associated with the pathogenesis of osteoarthritis (OA), the mRNA profile of m6A modification in OA remains unknown. Therefore, our study aimed to identify common m6A features and novel m6A-related therapeutic targets in OA. In the present study, we identified 3962 differentially methylated genes (DMGs) and 2048 differentially expressed genes (DEGs) using methylated RNA immunoprecipitation next-generation sequencing (MeRIP-seq) and RNA-sequencing. A co-expression analysis of DMGs and DEGs showed that the expression of 805 genes was significantly affected by m6A methylation. Specifically, we obtained 28 hypermethylated and upregulated genes, 657 hypermethylated and downregulated genes, 102 hypomethylated and upregulated genes, and 18 hypomethylated and downregulated genes. The differential gene expression analysis based on GSE114007 revealed 2770 DEGs. The Weighted Gene Co-expression Network Analysis (WGCNA) based on GSE114007 identified 134 OA-related genes. By taking the intersection of these results, ten novel aberrantly expressed, m6A-modified and OA-related key genes were identified, including SKP2, SULF1, TNC, ZFP36, CEBPB, BHLHE41, SOX9, VEGFA, MKNK2 and TUBB4B. The present study may provide valuable insight into identifying m6A-related pharmacological targets in OA.
Keywords: WGCNA; m6A methylation; merip-seq; methylome profile; osteoarthritis.
Copyright © 2023 Yu, Lu, Li and Xu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Synchronous profiling of mRNA N6-methyladenosine modifications and mRNA expression in high-grade serous ovarian cancer: a pilot study.Sci Rep. 2024 May 7;14(1):10427. doi: 10.1038/s41598-024-60975-x. Sci Rep. 2024. PMID: 38714753 Free PMC article.
-
Comprehensive Analysis of the Transcriptome-Wide m6A Methylome in Pterygium by MeRIP Sequencing.Front Cell Dev Biol. 2021 Jun 25;9:670528. doi: 10.3389/fcell.2021.670528. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34249924 Free PMC article.
-
DNER and GNL2 are differentially m6A methylated in periodontitis in comparison with periodontal health revealed by m6A microarray of human gingival tissue and transcriptomic analysis.J Periodontal Res. 2023 Jun;58(3):529-543. doi: 10.1111/jre.13117. Epub 2023 Mar 20. J Periodontal Res. 2023. PMID: 36941720
-
Regulatory Role of N6-Methyladenosine (m6A) Modification in Osteoarthritis.Front Cell Dev Biol. 2022 Jun 30;10:946219. doi: 10.3389/fcell.2022.946219. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35846376 Free PMC article. Review.
-
Novel insights into the interaction between N6-methyladenosine methylation and noncoding RNAs in musculoskeletal disorders.Cell Prolif. 2022 Oct;55(10):e13294. doi: 10.1111/cpr.13294. Epub 2022 Jun 23. Cell Prolif. 2022. PMID: 35735243 Free PMC article. Review.
Cited by
-
Elucidating the role of ubiquitination and deubiquitination in osteoarthritis progression.Front Immunol. 2023 Jun 9;14:1217466. doi: 10.3389/fimmu.2023.1217466. eCollection 2023. Front Immunol. 2023. PMID: 37359559 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

