Nonessential tRNA and rRNA modifications impact the bacterial response to sub-MIC antibiotic stress
- PMID: 37223353
- PMCID: PMC10117853
- DOI: 10.1093/femsml/uqac019
Nonessential tRNA and rRNA modifications impact the bacterial response to sub-MIC antibiotic stress
Abstract
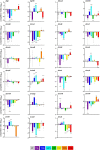
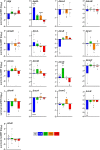
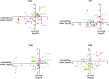
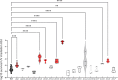
Antimicrobial resistance develops as a major problem in infectious diseases treatment. While antibiotic resistance mechanisms are usually studied using lethal antibiotic doses, lower doses allowing bacterial growth are now considered as factors influencing the development and selection of resistance. Starting with a high-density Tn insertion library in Vibrio cholerae and following its evolution by TN-seq in the presence of subinhibitory concentrations of antibiotics, we discovered that RNA modification genes can have opposite fates, being selected or counter-selected. We, thus have undertaken the phenotypic characterization of 23 transfer RNA (tRNA) and ribosomal RNA (rRNA) modifications deletion mutants, for which growth is globally not affected in the absence of stress. We uncover a specific involvement of different RNA modification genes in the response to aminoglycosides (tobramycin and gentamicin), fluoroquinolones (ciprofloxacin), β-lactams (carbenicillin), chloramphenicol, and trimethoprim. Our results identify t/rRNA modification genes, not previously associated to any antibiotic resistance phenotype, as important factors affecting the bacterial response to low doses of antibiotics from different families. This suggests differential translation and codon decoding as critical factors involved in the bacterial response to stress.
Keywords: RNA modifications; Vibrio cholerae; antibiotic resistance; bacterial stress responses; differential translation; sub-MIC antibiotics.
© The Author(s) 2022. Published by Oxford University Press on behalf of FEMS.
Conflict of interest statement
None declared.
Figures


Similar articles
-
RadD Contributes to R-Loop Avoidance in Sub-MIC Tobramycin.mBio. 2019 Jul 2;10(4):e01173-19. doi: 10.1128/mBio.01173-19. mBio. 2019. PMID: 31266870 Free PMC article.
-
Multidrug Adaptive Resistance of Pseudomonas aeruginosa Swarming Cells.Antimicrob Agents Chemother. 2020 Feb 21;64(3):e01999-19. doi: 10.1128/AAC.01999-19. Print 2020 Feb 21. Antimicrob Agents Chemother. 2020. PMID: 31844008 Free PMC article.
-
Low Ciprofloxacin Concentrations Select Multidrug-Resistant Mutants Overproducing Efflux Pumps in Clinical Isolates of Pseudomonas aeruginosa.Microbiol Spectr. 2022 Oct 26;10(5):e0072322. doi: 10.1128/spectrum.00723-22. Epub 2022 Aug 24. Microbiol Spectr. 2022. PMID: 36000896 Free PMC article.
-
Effect of subinhibitory antimicrobial concentrations (sub-MICs) on in-vitro bacterial adherence to uroepithelial cells.J Antimicrob Chemother. 1992 Jun;29(6):617-27. doi: 10.1093/jac/29.6.617. J Antimicrob Chemother. 1992. PMID: 1506346 Review.
-
In nosocomial pneumonia, optimizing antibiotics other than aminoglycosides is a more important determinant of successful clinical outcome, and a better means of avoiding resistance.Semin Respir Infect. 1997 Dec;12(4):278-93. Semin Respir Infect. 1997. PMID: 9436955 Review.
Cited by
-
What's the Matter with MICs: Bacterial Nutrition, Limiting Resources, and Antibiotic Pharmacodynamics.Microbiol Spectr. 2023 Jun 15;11(3):e0409122. doi: 10.1128/spectrum.04091-22. Epub 2023 May 3. Microbiol Spectr. 2023. PMID: 37130356 Free PMC article.
-
Biosynthesis and function of 7-deazaguanine derivatives in bacteria and phages.Microbiol Mol Biol Rev. 2024 Mar 27;88(1):e0019923. doi: 10.1128/mmbr.00199-23. Epub 2024 Feb 29. Microbiol Mol Biol Rev. 2024. PMID: 38421302 Free PMC article. Review.
-
Bacteria exposed to antiviral drugs develop antibiotic cross-resistance and unique resistance profiles.Commun Biol. 2023 Aug 12;6(1):837. doi: 10.1038/s42003-023-05177-3. Commun Biol. 2023. PMID: 37573457 Free PMC article.
-
RNA Post-transcriptional Modifications of an Early-Stage Large-Subunit Ribosomal Intermediate.Biochemistry. 2023 Oct 17;62(20):2908-2915. doi: 10.1021/acs.biochem.3c00291. Epub 2023 Sep 26. Biochemistry. 2023. PMID: 37751522 Free PMC article.
-
Two complete 1918 influenza A/H1N1 pandemic virus genomes characterized by next-generation sequencing using RNA isolated from formalin-fixed, paraffin-embedded autopsy lung tissue samples along with evidence of secondary bacterial co-infection.mBio. 2024 Mar 13;15(3):e0321823. doi: 10.1128/mbio.03218-23. Epub 2024 Feb 13. mBio. 2024. PMID: 38349163 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
