RNA-Chrom: a manually curated analytical database of RNA-chromatin interactome
- PMID: 37221043
- PMCID: PMC10205464
- DOI: 10.1093/database/baad025
RNA-Chrom: a manually curated analytical database of RNA-chromatin interactome
Erratum in
-
Correction to: RNA-Chrom: a manually curated analytical database of RNA-chromatin interactome.Database (Oxford). 2023 Jul 1;2023:baad052. doi: 10.1093/database/baad052. Database (Oxford). 2023. PMID: 37410919 Free PMC article. No abstract available.
Abstract
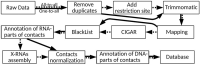
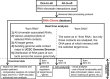
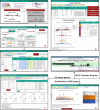
Every year there is more and more evidence that non-coding RNAs play an important role in biological processes affecting various levels of organization of living systems: from the cellular (regulation of gene expression, remodeling and maintenance of chromatin structure, co-transcriptional suppression of transposons, splicing, post-transcriptional RNA modifications, etc.) to cell populations and even organismal ones (development, aging, cancer, cardiovascular and many other diseases). The development and creation of mutually complementary databases that will aggregate, unify and structure different types of data can help to reach the system level of studying non-coding RNAs. Here we present the RNA-Chrom manually curated analytical database, which contains the coordinates of billions of contacts of thousands of human and mouse RNAs with chromatin. Through the user-friendly web interface (https://rnachrom2.bioinf.fbb.msu.ru/), two approaches to the analysis of the RNA-chromatin interactome were implemented. Firstly, to find out whether the RNA of interest to a user contacts with chromatin, and if so, with which genes or DNA loci? Secondly, to find out which RNAs are in contact with the DNA locus of interest to a user (and probably participate in its regulation), and if there are such, what is the nature of their interaction? For a more detailed study of contact maps and their comparison with other data, the web interface allows a user to view them in the UCSC Genome Browser. Database URL https://genome.ucsc.edu/.
© The Author(s) 2023. Published by Oxford University Press.
Figures



Similar articles
-
LnChrom: a resource of experimentally validated lncRNA-chromatin interactions in human and mouse.Database (Oxford). 2018 Jan 1;2018:bay039. doi: 10.1093/database/bay039. Database (Oxford). 2018. PMID: 29788225 Free PMC article.
-
Mapping Transcriptome-Wide and Genome-Wide RNA-DNA Contacts with Chromatin-Associated RNA Sequencing (ChAR-seq).Methods Mol Biol. 2020;2161:115-142. doi: 10.1007/978-1-0716-0680-3_10. Methods Mol Biol. 2020. PMID: 32681510 Free PMC article.
-
DASHR 2.0: integrated database of human small non-coding RNA genes and mature products.Bioinformatics. 2019 Mar 15;35(6):1033-1039. doi: 10.1093/bioinformatics/bty709. Bioinformatics. 2019. PMID: 30668832 Free PMC article.
-
Epigenomics in stress tolerance of plants under the climate change.Mol Biol Rep. 2023 Jul;50(7):6201-6216. doi: 10.1007/s11033-023-08539-6. Epub 2023 Jun 9. Mol Biol Rep. 2023. PMID: 37294468 Review.
-
RNA Epigenetics: Fine-Tuning Chromatin Plasticity and Transcriptional Regulation, and the Implications in Human Diseases.Genes (Basel). 2021 Apr 22;12(5):627. doi: 10.3390/genes12050627. Genes (Basel). 2021. PMID: 33922187 Free PMC article. Review.
Cited by
-
Functional identification of cis-regulatory long noncoding RNAs at controlled false discovery rates.Nucleic Acids Res. 2024 Apr 12;52(6):2821-2835. doi: 10.1093/nar/gkae075. Nucleic Acids Res. 2024. PMID: 38348970 Free PMC article.
-
BaRDIC: robust peak calling for RNA-DNA interaction data.NAR Genom Bioinform. 2024 May 20;6(2):lqae054. doi: 10.1093/nargab/lqae054. eCollection 2024 Jun. NAR Genom Bioinform. 2024. PMID: 38774512 Free PMC article.
-
Exploring the landscape of tools and resources for the analysis of long non-coding RNAs.Comput Struct Biotechnol J. 2023 Sep 29;21:4706-4716. doi: 10.1016/j.csbj.2023.09.041. eCollection 2023. Comput Struct Biotechnol J. 2023. PMID: 37841333 Free PMC article. Review.
-
Annotation of nuclear lncRNAs based on chromatin interactions.PLoS One. 2024 May 6;19(5):e0295971. doi: 10.1371/journal.pone.0295971. eCollection 2024. PLoS One. 2024. PMID: 38709794 Free PMC article.
References
-
- Brockdorff N., Ashworth A., Kay G.F.. et al. (1992) The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell, 71, 515–526. - PubMed
-
- Ryabykh G.K., Mylarshchikov D.E., Kuznetsov S.V.. et al. (2022) RNA-chromatin interactome: what? where? when?. Mol. Biol., 56, 210–228. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

