Vaccinia E5 is a major inhibitor of the DNA sensor cGAS
- PMID: 37217469
- PMCID: PMC10201048
- DOI: 10.1038/s41467-023-38514-5
Vaccinia E5 is a major inhibitor of the DNA sensor cGAS
Abstract
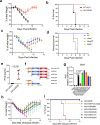
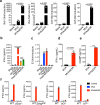
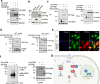
The DNA sensor cyclic GMP-AMP synthase (cGAS) is critical in host antiviral immunity. Vaccinia virus (VACV) is a large cytoplasmic DNA virus that belongs to the poxvirus family. How vaccinia virus antagonizes the cGAS-mediated cytosolic DNA-sensing pathway is not well understood. In this study, we screened 80 vaccinia genes to identify potential viral inhibitors of the cGAS/Stimulator of interferon gene (STING) pathway. We discovered that vaccinia E5 is a virulence factor and a major inhibitor of cGAS. E5 is responsible for abolishing cGAMP production during vaccinia virus (Western Reserve strain) infection of dendritic cells. E5 localizes to the cytoplasm and nucleus of infected cells. Cytosolic E5 triggers ubiquitination of cGAS and proteasome-dependent degradation via interacting with cGAS. Deleting the E5R gene from the Modified vaccinia virus Ankara (MVA) genome strongly induces type I IFN production by dendritic cells (DCs) and promotes DC maturation, and thereby improves antigen-specific T cell responses.
© 2023. The Author(s).
Conflict of interest statement
Memorial Sloan Kettering Cancer Center filed a patent application for the discovery of vaccinia viral inhibitors of the cytosolic DNA-sensing pathway and its use for improving MVA and vaccinia as oncolytic agents and vaccine vectors. The patent has been licensed to IMVAQ Therapeutics. L.D. and N.Y. are co-founders of IMVAQ Therapeutics. The remaining authors declare no competing interests.
Figures



Similar articles
-
Type I interferon-dependent CCL4 is induced by a cGAS/STING pathway that bypasses viral inhibition and protects infected tissue, independent of viral burden.PLoS Pathog. 2019 Oct 11;15(10):e1007778. doi: 10.1371/journal.ppat.1007778. eCollection 2019 Oct. PLoS Pathog. 2019. PMID: 31603920 Free PMC article.
-
cGAS exacerbates Schistosoma japonicum infection in a STING-type I IFN-dependent and independent manner.PLoS Pathog. 2022 Feb 2;18(2):e1010233. doi: 10.1371/journal.ppat.1010233. eCollection 2022 Feb. PLoS Pathog. 2022. PMID: 35108342 Free PMC article.
-
Efficient Induction of Cytotoxic T Cells by Viral Vector Vaccination Requires STING-Dependent DC Functions.Front Immunol. 2020 Jul 16;11:1458. doi: 10.3389/fimmu.2020.01458. eCollection 2020. Front Immunol. 2020. PMID: 32765505 Free PMC article.
-
cGAS/cGAMP/STING signal propagation in the tumor microenvironment: Key role for myeloid cells in antitumor immunity.Radiother Oncol. 2022 Sep;174:158-167. doi: 10.1016/j.radonc.2022.07.014. Epub 2022 Jul 20. Radiother Oncol. 2022. PMID: 35870728 Review.
-
Intrinsic strategies for the evasion of cGAS-STING signaling-mediated immune surveillance in human cancer: How therapy can overcome them.Pharmacol Res. 2021 Apr;166:105514. doi: 10.1016/j.phrs.2021.105514. Epub 2021 Feb 23. Pharmacol Res. 2021. PMID: 33631336 Review.
Cited by
-
Oncolytic vaccinia virus and cancer immunotherapy.Front Immunol. 2024 Jan 12;14:1324744. doi: 10.3389/fimmu.2023.1324744. eCollection 2023. Front Immunol. 2024. PMID: 38283361 Free PMC article. Review.
-
OX40L-expressing recombinant modified vaccinia virus Ankara induces potent antitumor immunity via reprogramming Tregs.J Exp Med. 2023 Aug 7;220(8):e20221166. doi: 10.1084/jem.20221166. Epub 2023 May 5. J Exp Med. 2023. PMID: 37145142 Free PMC article.
-
The poxvirus F17 protein counteracts mitochondrially orchestrated antiviral responses.Nat Commun. 2023 Nov 30;14(1):7889. doi: 10.1038/s41467-023-43635-y. Nat Commun. 2023. PMID: 38036506 Free PMC article.
-
Monkeypox virus: The changing facets of a zoonotic pathogen.Infect Genet Evol. 2022 Nov;105:105372. doi: 10.1016/j.meegid.2022.105372. Epub 2022 Oct 4. Infect Genet Evol. 2022. PMID: 36202208 Free PMC article. Review.
-
Microtubule disruption synergizes with STING signaling to show potent and broad-spectrum antiviral activity.PLoS Pathog. 2024 Feb 26;20(2):e1012048. doi: 10.1371/journal.ppat.1012048. eCollection 2024 Feb. PLoS Pathog. 2024. PMID: 38408104 Free PMC article.
References
-
- Ablasser, A. & Chen, Z. J. cGAS in action: expanding roles in immunity and inflammation. Science10.1126/science.aat8657 (2019). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

