Asymmetric conformations of cleaved HIV-1 envelope glycoprotein trimers in styrene-maleic acid lipid nanoparticles
- PMID: 37202420
- PMCID: PMC10195785
- DOI: 10.1038/s42003-023-04916-w
Asymmetric conformations of cleaved HIV-1 envelope glycoprotein trimers in styrene-maleic acid lipid nanoparticles
Abstract
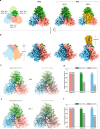
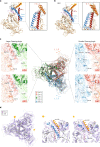
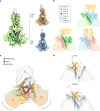
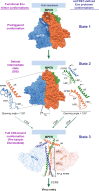
During virus entry, the pretriggered human immunodeficiency virus (HIV-1) envelope glycoprotein (Env) trimer initially transits into a default intermediate state (DIS) that remains structurally uncharacterized. Here, we present cryo-EM structures at near-atomic resolution of two cleaved full-length HIV-1 Env trimers purified from cell membranes in styrene-maleic acid lipid nanoparticles without antibodies or receptors. The cleaved Env trimers exhibited tighter subunit packing than uncleaved trimers. Cleaved and uncleaved Env trimers assumed remarkably consistent yet distinct asymmetric conformations, with one smaller and two larger opening angles. Breaking conformational symmetry is allosterically coupled with dynamic helical transformations of the gp41 N-terminal heptad repeat (HR1N) regions in two protomers and with trimer tilting in the membrane. The broken symmetry of the DIS potentially assists Env binding to two CD4 receptors-while resisting antibody binding-and promotes extension of the gp41 HR1 helical coiled-coil, which relocates the fusion peptide closer to the target cell membrane.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Asymmetric Structures and Conformational Plasticity of the Uncleaved Full-Length Human Immunodeficiency Virus Envelope Glycoprotein Trimer.J Virol. 2021 Nov 23;95(24):e0052921. doi: 10.1128/JVI.00529-21. Epub 2021 Sep 22. J Virol. 2021. PMID: 34549974 Free PMC article.
-
Conformational Differences between Functional Human Immunodeficiency Virus Envelope Glycoprotein Trimers and Stabilized Soluble Trimers.J Virol. 2019 Jan 17;93(3):e01709-18. doi: 10.1128/JVI.01709-18. Print 2019 Feb 1. J Virol. 2019. PMID: 30429345 Free PMC article.
-
Conformations of Human Immunodeficiency Virus Envelope Glycoproteins in Detergents and Styrene-Maleic Acid Lipid Particles.J Virol. 2023 Jun 29;97(6):e0032723. doi: 10.1128/jvi.00327-23. Epub 2023 May 31. J Virol. 2023. PMID: 37255444 Free PMC article.
-
The HIV-1 envelope glycoprotein structure: nailing down a moving target.Immunol Rev. 2017 Jan;275(1):21-32. doi: 10.1111/imr.12507. Immunol Rev. 2017. PMID: 28133813 Free PMC article. Review.
-
HIV-1 gp41: mediator of fusion and target for inhibition.AIDS Rev. 2003 Oct-Dec;5(4):214-21. AIDS Rev. 2003. PMID: 15012000 Review.
Cited by
-
Conformations of membrane human immunodeficiency virus (HIV-1) envelope glycoproteins solubilized in Amphipol A18 lipid-nanodiscs.J Virol. 2024 Oct 22;98(10):e0063124. doi: 10.1128/jvi.00631-24. Epub 2024 Sep 9. J Virol. 2024. PMID: 39248459
-
Alternative substitutions of N332 in HIV-1AD8 gp120 differentially affect envelope glycoprotein function and viral sensitivity to broadly neutralizing antibodies targeting the V3-glycan.mBio. 2024 Apr 10;15(4):e0268623. doi: 10.1128/mbio.02686-23. Epub 2024 Mar 12. mBio. 2024. PMID: 38470051 Free PMC article.
-
Stoichiometry of HIV-1 Envelope Glycoprotein Protomers with Changes That Stabilize or Destabilize the Pretriggered Conformation.bioRxiv [Preprint]. 2024 Oct 25:2024.10.25.620268. doi: 10.1101/2024.10.25.620268. bioRxiv. 2024. PMID: 39484577 Free PMC article. Preprint.
-
Progress with induction of HIV broadly neutralizing antibodies in the Duke Consortia for HIV/AIDS Vaccine Development.Curr Opin HIV AIDS. 2023 Nov 1;18(6):300-308. doi: 10.1097/COH.0000000000000820. Epub 2023 Sep 25. Curr Opin HIV AIDS. 2023. PMID: 37751363 Free PMC article. Review.
-
Alterations in gp120 glycans or the gp41 fusion peptide-proximal region modulate the stability of the human immunodeficiency virus (HIV-1) envelope glycoprotein pretriggered conformation.J Virol. 2023 Sep 28;97(9):e0059223. doi: 10.1128/jvi.00592-23. Epub 2023 Sep 11. J Virol. 2023. PMID: 37696048 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

