SESN1 is a FOXO3 effector that counteracts human skeletal muscle ageing
- PMID: 37199024
- PMCID: PMC10212707
- DOI: 10.1111/cpr.13455
SESN1 is a FOXO3 effector that counteracts human skeletal muscle ageing
Abstract
Sarcopenia, a skeletal muscle disorder in which loss of muscle mass and function progresses with age, is associated with increased overall frailty, risk of falling and mortality in the elders. Here, we reveal that SESN1 safeguards skeletal muscle from ageing downstream of the longevity gene FOXO3, which we recently reported is a geroprotector in primate skeletal muscle. Knockdown of SESN1 mimicked the human myotube ageing phenotypes observed in the FOXO3-deficient human myotubes, whereas genetic activation of SESN1 alleviated human myotube senescence. Of note, SESN1 was identified as a protective secretory factor against muscle atrophy. Administration of recombinant SESN1 protein attenuated senescence of human myotubes in vitro and facilitated muscle regeneration in vivo. Altogether, we unveil a key role of SESN1 downstream of FOXO3 in protecting skeletal muscle from ageing, providing diagnostic biomarkers and intervention strategies for counteracting skeletal muscle ageing and related diseases.
© 2023 The Authors. Cell Proliferation published by Beijing Institute for Stem Cell and Regenerative Medicine and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

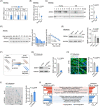

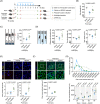

Similar articles
-
Single-nucleus profiling unveils a geroprotective role of the FOXO3 in primate skeletal muscle aging.Protein Cell. 2023 Jun 28;14(7):497-512. doi: 10.1093/procel/pwac061. Protein Cell. 2023. PMID: 36921027 Free PMC article.
-
Foxo3 Knockdown Mediates Decline of Myod1 and Myog Reducing Myoblast Conversion to Myotubes.Cells. 2023 Aug 29;12(17):2167. doi: 10.3390/cells12172167. Cells. 2023. PMID: 37681900 Free PMC article.
-
Regulation of muscle atrophy-related genes by the opposing transcriptional activities of ZEB1/CtBP and FOXO3.Nucleic Acids Res. 2018 Nov 16;46(20):10697-10708. doi: 10.1093/nar/gky835. Nucleic Acids Res. 2018. PMID: 30304480 Free PMC article.
-
Therapeutic Consequences of Targeting the IGF-1/PI3K/AKT/FOXO3 Axis in Sarcopenia: A Narrative Review.Cells. 2023 Dec 7;12(24):2787. doi: 10.3390/cells12242787. Cells. 2023. PMID: 38132107 Free PMC article. Review.
-
Recent advances in understanding the role of FOXO3.F1000Res. 2018 Aug 31;7:F1000 Faculty Rev-1372. doi: 10.12688/f1000research.15258.1. eCollection 2018. F1000Res. 2018. PMID: 30228872 Free PMC article. Review.
Cited by
-
Sestrin1, 2, and 3 are dispensable for female fertility in mice.J Ovarian Res. 2024 Feb 1;17(1):28. doi: 10.1186/s13048-024-01345-z. J Ovarian Res. 2024. PMID: 38297375 Free PMC article.
-
Microbiota-derived 3-phenylpropionic acid promotes myotube hypertrophy by Foxo3/NAD+ signaling pathway.Cell Biosci. 2024 May 15;14(1):62. doi: 10.1186/s13578-024-01244-2. Cell Biosci. 2024. PMID: 38750565 Free PMC article.
-
Exploring In Vivo Models of Musculoskeletal Frailty: A Comprehensive Systematic Review.Int J Mol Sci. 2023 Nov 29;24(23):16948. doi: 10.3390/ijms242316948. Int J Mol Sci. 2023. PMID: 38069274 Free PMC article. Review.
-
Degeneration Directory: a multi-omics web resource for degenerative diseases.Protein Cell. 2024 May 7;15(5):385-392. doi: 10.1093/procel/pwad066. Protein Cell. 2024. PMID: 38153694 Free PMC article. No abstract available.
-
Beyond the bulk: overview and novel insights into the dynamics of muscle satellite cells during muscle regeneration.Inflamm Regen. 2024 Sep 26;44(1):39. doi: 10.1186/s41232-024-00354-1. Inflamm Regen. 2024. PMID: 39327631 Free PMC article. Review.
References
-
- Zou X, Dai X, Mentis A.‐F. A, et al. From monkey single‐cell atlases into a broader biomedical perspective. Life Med. 2022;1(3):254‐257.
-
- Cruz‐Jentoft AJ, Sayer AA. Sarcopenia. Lancet. 2019;393(10191):2636‐2646. - PubMed
-
- Wang Y, Shyh‐Chang N. Sphingolipids mediate lipotoxicity in muscular dystrophies. Life Med. 2022;1(3):273‐275.
-
- Woo J. Sarcopenia. Clin Geriatr Med. 2017;33(3):305‐314. - PubMed
MeSH terms
Substances
Grants and funding
- 2022YFA1103700/National Key Research and Development Program of China
- XDA16000000/Strategic Priority Research Program of the Chinese Academy of Sciences
- 2020YFA0804000/National Key Research and Development Program of China
- 2018YFC2000100/National Key Research and Development Program of China
- STI2030-Major Projects-2021ZD0202400/National Key Research and Development Program of China
- 2020YFA0112200/National Key Research and Development Program of China
- 2018YFA0107203/National Key Research and Development Program of China
- 2021YFF1201005/National Key Research and Development Program of China
- 2021YFE0111800/National Key Research and Development Program of China
- 82071588/National Natural Science Foundation of China
- 81921006/National Natural Science Foundation of China
- 82125011/National Natural Science Foundation of China
- 92149301/National Natural Science Foundation of China
- 92168201/National Natural Science Foundation of China
- 92049116/National Natural Science Foundation of China
- 32121001/National Natural Science Foundation of China
- 82192863/National Natural Science Foundation of China
- 91949209/National Natural Science Foundation of China
- 92049304/National Natural Science Foundation of China
- 82122024/National Natural Science Foundation of China
- 82001477/National Natural Science Foundation of China
- 32000500/National Natural Science Foundation of China
- 82271600/National Natural Science Foundation of China
- 82201714/National Natural Science Foundation of China
- 91849132/National Natural Science Foundation of China
- 82225019/National Natural Science Foundation of China
- Z190019/Program of the Beijing Natural Science Foundation
- GJTD-2019-06/K. C. Wong Education Foundation
- GJTD-2019-08/K. C. Wong Education Foundation
- 2021-8/Beijing Medical Research
- 11000022T000000461062/the Pilot Project for Public Welfare Development and Reform of Beijing-affiliated Medical Research Institutes
- YESS20200012/Young Elite Scientists Sponsorship Program by CAST
- YESS20210002/Young Elite Scientists Sponsorship Program by CAST
- YSBR-076/CAS Project for Young Scientists in Basic Research
- YSBR-012/CAS Project for Young Scientists in Basic Research
- E1CAZW0401/Youth Innovation Promotion Association of CAS
- 2020085/Youth Innovation Promotion Association of CAS
- 2022083/Youth Innovation Promotion Association of CAS
- CAS-WX2022SDC-XK14/Informatization Plan of Chinese Academy of Sciences
- CAS-WX2021SF-0301/Informatization Plan of Chinese Academy of Sciences
- CAS-WX2021SF-0101/Informatization Plan of Chinese Academy of Sciences
- 2021-1045/Tencent Foundation
- 2022M712216/The Fellowship of China Postdoctoral Science Foundation
- QML20230806/Beijing Hospitals Authority Youth Programme
- 2021-I2M-1-050/CAMS Innovation Fund for Medical Sciences
- 202001AY070001-011/Yunnan Priority Union Foundation
LinkOut - more resources
Full Text Sources
Research Materials

