Genome-Wide Mapping Implicates 5-Hydroxymethylcytosines in Diabetes Mellitus and Alzheimer's Disease
- PMID: 37182870
- PMCID: PMC10490934
- DOI: 10.3233/JAD-221113
Genome-Wide Mapping Implicates 5-Hydroxymethylcytosines in Diabetes Mellitus and Alzheimer's Disease
Abstract
Background: Diabetes mellitus (DM) is a recognized risk factor for dementia. Because DM is a potentially modifiable condition, greater understanding of the mechanisms linking DM to the clinical expression of Alzheimer's disease dementia may provide insights into much needed dementia therapeutics.
Objective: In this feasibility study, we investigated DM as a dementia risk factor by examining genome-wide distributions of the epigenetic DNA modification 5-hydroxymethylcytosine (5hmC).
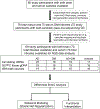
Methods: We obtained biologic samples from the Rush Memory and Aging Project and used the highly sensitive 5hmC-Seal technique to perform genome-wide profiling of 5hmC in circulating cell-free DNA (cfDNA) from antemortem serum samples and in genomic DNA from postmortem prefrontal cortex brain tissue from 80 individuals across four groups: Alzheimer's disease neuropathologically defined (AD), DM clinically defined, AD with DM, and individuals with neither disease (controls).
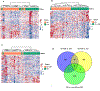
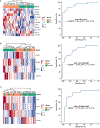
Results: Distinct 5hmC signatures and biological pathways were enriched in persons with both AD and DM versus AD alone, DM alone, or controls, including genes inhibited by EGFR signaling in oligodendroglia and those activated by constitutive RHOA. We also demonstrate the potential diagnostic value of 5hmC profiling in circulating cfDNA. Specifically, an 11-gene weighted model distinguished AD from non-AD/non-DM controls (AUC = 91.8%; 95% CI, 82.9-100.0%), while a 4-gene model distinguished DM-associated AD from AD alone (AUC = 87.9%; 95% CI, 77.5-98.3%).
Conclusion: We demonstrate in this small sample, the feasibility of detecting and characterizing 5hmC in DM-associated AD and of using 5hmC information contained in circulating cfDNA to detect AD in high-risk individuals, such as those with diabetes.
Keywords: 5-hydroxymethylcytosine; Alzheimer’s disease; cell-free DNA; dementia; diabetes mellitus; epigenetics.
Conflict of interest statement
Competing interests
CH was the scientific founder of Epican Genetech, which obtained a license from the University of Chicago to develop the 5hmC-Seal technique for clinical applications. WZ was an advisor of Epican Genetech, and he received research support from the company.
Figures


Similar articles
-
5-Hydroxymethylcytosine Signatures in Circulating Cell-Free DNA as Diagnostic Biomarkers for Late-Onset Alzheimer's Disease.J Alzheimers Dis. 2022;85(2):573-585. doi: 10.3233/JAD-215217. J Alzheimers Dis. 2022. PMID: 34864677
-
5-Hydroxymethylcytosine signature in circulating cell-free DNA as a potential diagnostic factor for early-stage colorectal cancer and precancerous adenoma.Mol Oncol. 2021 Jan;15(1):138-150. doi: 10.1002/1878-0261.12833. Epub 2020 Nov 14. Mol Oncol. 2021. PMID: 33107199 Free PMC article.
-
Alterations of 5-hydroxymethylcytosines in circulating cell-free DNA reflect retinopathy in type 2 diabetes.Genomics. 2021 Jan;113(1 Pt 1):79-87. doi: 10.1016/j.ygeno.2020.11.014. Epub 2020 Nov 20. Genomics. 2021. PMID: 33221518
-
Epigenetics in Alzheimer's disease: a focus on DNA modifications.Curr Pharm Des. 2011;17(31):3398-412. doi: 10.2174/138161211798072544. Curr Pharm Des. 2011. PMID: 21902668 Review.
-
Genomic distribution and possible functions of DNA hydroxymethylation in the brain.Genomics. 2014 Nov;104(5):341-6. doi: 10.1016/j.ygeno.2014.08.020. Epub 2014 Sep 7. Genomics. 2014. PMID: 25205307 Review.
Cited by
-
Liquid Biopsy in Neuropsychiatric Disorders: A Step Closer to Precision Medicine.Mol Neurobiol. 2024 Sep 19. doi: 10.1007/s12035-024-04492-y. Online ahead of print. Mol Neurobiol. 2024. PMID: 39298102 Review.
-
A 5-Hydroxymethylcytosine-Based Noninvasive Model for Early Detection of Colorectal Carcinomas and Advanced Adenomas: The METHOD-2 Study.Clin Cancer Res. 2024 Aug 1;30(15):3337-3348. doi: 10.1158/1078-0432.CCR-24-0199. Clin Cancer Res. 2024. PMID: 38814264
-
PETCH-DB: a Portal for Exploring Tissue-specific and Complex disease-associated 5-Hydroxymethylcytosines.Database (Oxford). 2023 Jun 10;2023:baad042. doi: 10.1093/database/baad042. Database (Oxford). 2023. PMID: 37387524 Free PMC article.
-
Analysis of genome-wide 5-hydroxymethylation of blood samples stored in different anticoagulants: opportunities for the expansion of clinical resources for epigenetic research.Epigenetics. 2023 Dec;18(1):2271692. doi: 10.1080/15592294.2023.2271692. Epub 2023 Oct 29. Epigenetics. 2023. PMID: 37898998 Free PMC article.
References
-
- Blázquez E, Hurtado-Carneiro V, LeBaut-Ayuso Y, Velázquez E, García-García L, Gómez-Oliver F, Ruiz-Albusac JM, Ávila J, Pozo MÁ (2022) Significance of Brain Glucose Hypometabolism, Altered Insulin Signal Transduction, and Insulin Resistance in Several Neurological Diseases. Front Endocrinol (Lausanne) 13, 873301. - PMC - PubMed
-
- Arvanitakis Z, Wilson RS, Bienias JL, Evans DA, Bennett DA (2004) Diabetes mellitus and risk of Alzheimer disease and decline in cognitive function. Arch Neurol 61, 661–666. - PubMed
-
- Simó R, Ciudin A, Simó-Servat O, Hernández C (2017) Cognitive impairment and dementia: a new emerging complication of type 2 diabetes-The diabetologist’s perspective. Acta Diabetol 54, 417–424. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

