Genetic basis controlling rice plant architecture and its modification for breeding
- PMID: 37168811
- PMCID: PMC10165344
- DOI: 10.1270/jsbbs.22088
Genetic basis controlling rice plant architecture and its modification for breeding
Abstract
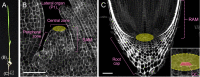
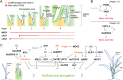
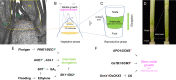
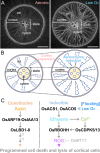
The shoot and root system architectures are fundamental for crop productivity. During the history of artificial selection of domestication and post-domestication breeding, the architecture of rice has significantly changed from its wild ancestor to fulfil requirements in agriculture. We review the recent studies on developmental biology in rice by focusing on components determining rice plant architecture; shoot meristems, leaves, tillers, stems, inflorescences and roots. We also highlight natural variations that affected these structures and were utilized in cultivars. Importantly, many core regulators identified from developmental mutants have been utilized in breeding as weak alleles moderately affecting these architectures. Given a surge of functional genomics and genome editing, the genetic mechanisms underlying the rice plant architecture discussed here will provide a theoretical basis to push breeding further forward not only in rice but also in other crops and their wild relatives.
Keywords: growth and development; inflorescence; leaf; root; shoot; stem.
Copyright © 2023 by JAPANESE SOCIETY OF BREEDING.
Figures





Similar articles
-
Genetic Regulation of Shoot Architecture.Annu Rev Plant Biol. 2018 Apr 29;69:437-468. doi: 10.1146/annurev-arplant-042817-040422. Epub 2018 Mar 19. Annu Rev Plant Biol. 2018. PMID: 29553800 Review.
-
Genetic insights into the modification of the pre-fertilization mechanisms during plant domestication.J Exp Bot. 2019 Jun 1;70(11):3007-3019. doi: 10.1093/jxb/erz231. J Exp Bot. 2019. PMID: 31152173 Review.
-
Genetic control of inflorescence architecture during rice domestication.Nat Commun. 2013;4:2200. doi: 10.1038/ncomms3200. Nat Commun. 2013. PMID: 23884108 Free PMC article.
-
Genome editing as a tool to achieve the crop ideotype and de novo domestication of wild relatives: Case study in tomato.Plant Sci. 2017 Mar;256:120-130. doi: 10.1016/j.plantsci.2016.12.012. Epub 2016 Dec 28. Plant Sci. 2017. PMID: 28167025 Review.
-
Enhancement of Plant Productivity in the Post-Genomics Era.Curr Genomics. 2016 Aug;17(4):295-6. doi: 10.2174/138920291704160607182507. Curr Genomics. 2016. PMID: 27499678 Free PMC article.
Cited by
-
Genomic introgressions from African rice (Oryza glaberrima) in Asian rice (O. sativa) lead to the identification of key QTLs for panicle architecture.BMC Genomics. 2023 Oct 4;24(1):587. doi: 10.1186/s12864-023-09695-6. BMC Genomics. 2023. PMID: 37794325 Free PMC article.
-
Possible Roles of Carbohydrate Management and Cytokinin in the Process of Defoliation-Regrowth Cycles in Rice.Int J Mol Sci. 2024 May 7;25(10):5070. doi: 10.3390/ijms25105070. Int J Mol Sci. 2024. PMID: 38791109 Free PMC article.
-
The Function of Florigen in the Vegetative-to-Reproductive Phase Transition in and around the Shoot Apical Meristem.Plant Cell Physiol. 2024 Apr 16;65(3):322-337. doi: 10.1093/pcp/pcae001. Plant Cell Physiol. 2024. PMID: 38179836 Free PMC article. Review.
-
Age-dependent analysis dissects the stepwise control of auxin-mediated lateral root development in rice.Plant Physiol. 2024 Jan 31;194(2):819-831. doi: 10.1093/plphys/kiad548. Plant Physiol. 2024. PMID: 37831077 Free PMC article.
-
Heat shock-inducible clonal analysis reveals the stepwise establishment of cell fate in the rice stem.Plant Cell. 2023 Nov 30;35(12):4366-4382. doi: 10.1093/plcell/koad241. Plant Cell. 2023. PMID: 37757885 Free PMC article.
References
-
- Abe, M., Kuroshita H., Umeda M., Itoh J. and Nagato Y. (2008) The rice FLATTENED SHOOT MERISTEM, encoding CAF-1 p150 subunit, is required for meristem maintenance by regulating the cell-cycle period. Dev Biol 319: 384–393. - PubMed
-
- Adedze, Y.M.N., Feng B., Shi L., Sheng Z., Tang S., Wei X. and Hu P. (2018) Further insight into the role of KAN1, a member of KANADI transcription factor family in rice. Plant Growth Regul 84: 237–248.
-
- Alvarez, J. and Smyth D.R. (1999) CRABS CLAW and SPATULA, two Arabidopsis genes that control carpel development in parallel with AGAMOUS. Development 126: 2377–2386. - PubMed

