Role of condensates in modulating DNA repair pathways and its implication for chemoresistance
- PMID: 37164156
- PMCID: PMC10318469
- DOI: 10.1016/j.jbc.2023.104800
Role of condensates in modulating DNA repair pathways and its implication for chemoresistance
Abstract
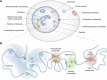
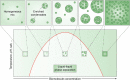
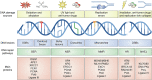
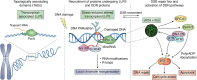
For cells, it is important to repair DNA damage, such as double-strand and single-strand DNA breaks, because unrepaired DNA can compromise genetic integrity, potentially leading to cell death or cancer. Cells have multiple DNA damage repair pathways that have been the subject of detailed genetic, biochemical, and structural studies. Recently, the scientific community has started to gain evidence that the repair of DNA double-strand breaks may occur within biomolecular condensates and that condensates may also contribute to DNA damage through concentrating genotoxic agents used to treat various cancers. Here, we summarize key features of biomolecular condensates and note where they have been implicated in the repair of DNA double-strand breaks. We also describe evidence suggesting that condensates may be involved in the repair of other types of DNA damage, including single-strand DNA breaks, nucleotide modifications (e.g., mismatch and oxidized bases), and bulky lesions, among others. Finally, we discuss old and new mysteries that could now be addressed considering the properties of condensates, including chemoresistance mechanisms.
Keywords: DNA damage response; chemoresistance; condensates; internal disordered regions; liquid-liquid phase separation.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
Similar articles
-
Single-cell microarray enables high-throughput evaluation of DNA double-strand breaks and DNA repair inhibitors.Cell Cycle. 2013 Mar 15;12(6):907-15. doi: 10.4161/cc.23880. Epub 2013 Feb 19. Cell Cycle. 2013. PMID: 23422001 Free PMC article.
-
The evolving complexity of DNA damage foci: RNA, condensates and chromatin in DNA double-strand break repair.DNA Repair (Amst). 2021 Sep;105:103170. doi: 10.1016/j.dnarep.2021.103170. Epub 2021 Jun 30. DNA Repair (Amst). 2021. PMID: 34256335 Free PMC article. Review.
-
A synthetic lethal therapeutic approach: poly(ADP) ribose polymerase inhibitors for the treatment of cancers deficient in DNA double-strand break repair.J Clin Oncol. 2008 Aug 1;26(22):3785-90. doi: 10.1200/JCO.2008.16.0812. Epub 2008 Jun 30. J Clin Oncol. 2008. PMID: 18591545 Review.
-
Repair Foci as Liquid Phase Separation: Evidence and Limitations.Genes (Basel). 2022 Oct 13;13(10):1846. doi: 10.3390/genes13101846. Genes (Basel). 2022. PMID: 36292731 Free PMC article. Review.
-
DNA repair targeted therapy: The past or future of cancer treatment?Pharmacol Ther. 2016 Apr;160:65-83. doi: 10.1016/j.pharmthera.2016.02.003. Epub 2016 Feb 16. Pharmacol Ther. 2016. PMID: 26896565 Free PMC article. Review.
Cited by
-
The strand exchange domain of tumor suppressor PALB2 is intrinsically disordered and promotes oligomerization-dependent DNA compaction.bioRxiv [Preprint]. 2024 May 29:2023.06.01.543259. doi: 10.1101/2023.06.01.543259. bioRxiv. 2024. Update in: iScience. 2024 Oct 28;27(12):111259. doi: 10.1016/j.isci.2024.111259 PMID: 37333393 Free PMC article. Updated. Preprint.
-
Membraneless organelles in health and disease: exploring the molecular basis, physiological roles and pathological implications.Signal Transduct Target Ther. 2024 Nov 18;9(1):305. doi: 10.1038/s41392-024-02013-w. Signal Transduct Target Ther. 2024. PMID: 39551864 Free PMC article. Review.
-
DNA Repair Protein XRCC1 Stimulates Activity of DNA Polymerase λ under Conditions of Microphase Separation.Int J Mol Sci. 2024 Jun 25;25(13):6927. doi: 10.3390/ijms25136927. Int J Mol Sci. 2024. PMID: 39000034 Free PMC article.
-
Revisiting Two Decades of Research Focused on Targeting APE1 for Cancer Therapy: The Pros and Cons.Cells. 2023 Jul 20;12(14):1895. doi: 10.3390/cells12141895. Cells. 2023. PMID: 37508559 Free PMC article. Review.
-
Divalent and multivalent cations control liquid-like assembly of poly(ADP-ribosyl)ated PARP1 into multimolecular associates in vitro.Commun Biol. 2024 Sep 15;7(1):1148. doi: 10.1038/s42003-024-06811-4. Commun Biol. 2024. PMID: 39278937 Free PMC article.
References
-
- Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

