Structural basis of DNA binding by the WhiB-like transcription factor WhiB3 in Mycobacterium tuberculosis
- PMID: 37142222
- PMCID: PMC10245118
- DOI: 10.1016/j.jbc.2023.104777
Structural basis of DNA binding by the WhiB-like transcription factor WhiB3 in Mycobacterium tuberculosis
Abstract
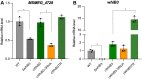
Mycobacterium tuberculosis (Mtb) WhiB3 is an iron-sulfur cluster-containing transcription factor belonging to a subclass of the WhiB-Like (Wbl) family that is widely distributed in the phylum Actinobacteria. WhiB3 plays a crucial role in the survival and pathogenesis of Mtb. It binds to the conserved region 4 of the principal sigma factor (σA4) in the RNA polymerase holoenzyme to regulate gene expression like other known Wbl proteins in Mtb. However, the structural basis of how WhiB3 coordinates with σA4 to bind DNA and regulate transcription is unclear. Here we determined crystal structures of the WhiB3:σA4 complex without and with DNA at 1.5 Å and 2.45 Å, respectively, to elucidate how WhiB3 interacts with DNA to regulate gene expression. These structures reveal that the WhiB3:σA4 complex shares a molecular interface similar to other structurally characterized Wbl proteins and also possesses a subclass-specific Arg-rich DNA-binding motif. We demonstrate that this newly defined Arg-rich motif is required for WhiB3 binding to DNA in vitro and transcriptional regulation in Mycobacterium smegmatis. Together, our study provides empirical evidence of how WhiB3 regulates gene expression in Mtb by partnering with σA4 and engaging with DNA via the subclass-specific structural motif, distinct from the modes of DNA interaction by WhiB1 and WhiB7.
Keywords: Wbl family; WhiB3; X-ray crystallography; iron–sulfur cluster; transcription factor.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare no conflict of interest with the contents of this article.
Figures

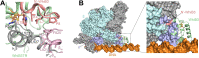



Similar articles
-
WhiB-like proteins: Diversity of structure, function and mechanism.Biochim Biophys Acta Mol Cell Res. 2024 Oct;1871(7):119787. doi: 10.1016/j.bbamcr.2024.119787. Epub 2024 Jun 13. Biochim Biophys Acta Mol Cell Res. 2024. PMID: 38879133 Review.
-
Structural insights into the functional divergence of WhiB-like proteins in Mycobacterium tuberculosis.Mol Cell. 2021 Jul 15;81(14):2887-2900.e5. doi: 10.1016/j.molcel.2021.06.002. Epub 2021 Jun 24. Mol Cell. 2021. PMID: 34171298 Free PMC article.
-
Mycobacterium tuberculosis WhiB3 interacts with RpoV to affect host survival but is dispensable for in vivo growth.Proc Natl Acad Sci U S A. 2002 Mar 5;99(5):3147-52. doi: 10.1073/pnas.052705399. Proc Natl Acad Sci U S A. 2002. PMID: 11880648 Free PMC article.
-
Structural basis for transcription initiation by bacterial ECF σ factors.Nat Commun. 2019 Mar 11;10(1):1153. doi: 10.1038/s41467-019-09096-y. Nat Commun. 2019. PMID: 30858373 Free PMC article.
-
Sigma factors of Mycobacterium tuberculosis.Tuber Lung Dis. 1997;78(3-4):175-83. doi: 10.1016/s0962-8479(97)90024-1. Tuber Lung Dis. 1997. PMID: 9713650 Review. No abstract available.
Cited by
-
WhiB-like proteins: Diversity of structure, function and mechanism.Biochim Biophys Acta Mol Cell Res. 2024 Oct;1871(7):119787. doi: 10.1016/j.bbamcr.2024.119787. Epub 2024 Jun 13. Biochim Biophys Acta Mol Cell Res. 2024. PMID: 38879133 Review.
-
Gene Regulatory Mechanism of Mycobacterium Tuberculosis during Dormancy.Curr Issues Mol Biol. 2024 Jun 11;46(6):5825-5844. doi: 10.3390/cimb46060348. Curr Issues Mol Biol. 2024. PMID: 38921019 Free PMC article. Review.
References
-
- Pai M., Kasaeva T., Swaminathan S. Covid-19's devastating effect on tuberculosis care - a path to recovery. N. Engl. J. Med. 2022;386:1490–1493. - PubMed
-
- Davis N.K., Chater K.F. The Streptomyces coelicolor whiB gene encodes a small transcription factor-like protein dispensable for growth but essential for sporulation. Mol. Gen. Genet. 1992;232:351–358. - PubMed
-
- Soliveri J.A., Gomez J., Bishai W.R., Chater K.F. Multiple paralogous genes related to the Streptomyces coelicolor developmental regulatory gene whiB are present in Streptomyces and other actinomycetes. Microbiology. 2000;146:333–343. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

