The CDR3 region as the major driver of TREM-1 interaction with its ligands, an in silico characterization
- PMID: 37122631
- PMCID: PMC10130352
- DOI: 10.1016/j.csbj.2023.04.008
The CDR3 region as the major driver of TREM-1 interaction with its ligands, an in silico characterization
Abstract
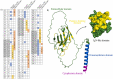
The triggering receptor expressed on myeloid cells-1 (TREM-1) is a pattern recognition receptor heavily investigated in infectious and non-infectious diseases. Because of its role in amplifying inflammation, TREM-1 has been explored as a diagnostic/prognostic biomarker. Further, as the receptor has been implicated in the pathophysiology of several diseases, therapies aiming at modulating its activity represent a promising strategy to constrain uncontrolled inflammatory or infectious diseases. Despite this, several aspects concerning its interaction with ligands and activation process, remain unclear. Although many molecules have been suggested as TREM-1 ligands, only five have been confirmed to interact with the receptor: actin, eCIRP, HMGB1, Hsp70 and PGLYRP1. However, the domains involved in the interaction between the receptor and these proteins are not clarified yet. Therefore, here we used in silico approaches to investigate the putative binding domains in the receptor, using hot spots analysis, molecular docking and molecular dynamics simulations between TREM-1 and its five known ligands. Our results indicated the complementarity-determining regions (CDRs) of the receptor as the main mediators of antigen recognition, especially the CDR3 loop. We believe that our study could be used as structural basis for the elucidation of TREM-1's recognition process, and may be useful for prospective in silico and biological investigations exploring the receptor in different contexts.
Keywords: Binding domain; CDR3; Pattern recognition receptor; TREM-1; TREM-1 ligands.
© 2023 Published by Elsevier B.V. on behalf of Research Network of Computational and Structural Biotechnology.
Conflict of interest statement
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Helioswilton Sales-Campos reports financial support was provided by Fundação de Apoio a Pesquisa (FUNAPE). Carolina Horta Andrade reports a relationship with Fundação de Amparo a Pesquisa do Estado de Goiás (FAPEG) that includes: funding grants. Bruno Junior Neves reports a relationship with Fundação de Amparo a Pesquisa do Estado de Goiás (FAPEG) that includes: funding grants. Carolina Horta Andrade reports a relationship with Conselho Nacional de Desenvolvimento Científico (CNPq) that includes: funding grants. Bruno Junior Neves reports a relationship with Conselho Nacional de Desenvolvimento Científico (CNPq) that includes: funding grants. Helioswilton Sales-Campos reports a relationship with Fundação de Amparo a Pesquisa (FUNAPE) that includes: funding grants. Bruno Junior Neves reports a relationship with Fundação de Amparo a Pesquisa (FUNAPE) that includes: funding grants.
Figures





Similar articles
-
Novel ligands and modulators of triggering receptor expressed on myeloid cells receptor family: 2015-2020 updates.Expert Opin Ther Pat. 2021 Jun;31(6):549-561. doi: 10.1080/13543776.2021.1883587. Epub 2021 Feb 8. Expert Opin Ther Pat. 2021. PMID: 33507843 Free PMC article. Review.
-
Extracellular CIRP as an endogenous TREM-1 ligand to fuel inflammation in sepsis.JCI Insight. 2020 Mar 12;5(5):e134172. doi: 10.1172/jci.insight.134172. JCI Insight. 2020. PMID: 32027618 Free PMC article.
-
TREM-1 and its potential ligands in non-infectious diseases: from biology to clinical perspectives.Pharmacol Ther. 2017 Sep;177:81-95. doi: 10.1016/j.pharmthera.2017.02.043. Epub 2017 Feb 27. Pharmacol Ther. 2017. PMID: 28245991 Review.
-
Triggering receptor expressed on myeloid cells-1 (TREM-1) as a therapeutic target in infectious and noninfectious disease: a critical review.Int Rev Immunol. 2020;39(4):188-202. doi: 10.1080/08830185.2020.1762597. Epub 2020 May 7. Int Rev Immunol. 2020. PMID: 32379561 Review.
-
N-Terminal Peptide of PGLYRP1/Tag7 Is a Novel Ligand for TREM-1 Receptor.Int J Mol Sci. 2022 May 20;23(10):5752. doi: 10.3390/ijms23105752. Int J Mol Sci. 2022. PMID: 35628562 Free PMC article.
Cited by
-
The orchestrating role of deteriorating neurons and TREM-1 in crosstalk with SYK in Alzheimer's disease progression and neuroinflammation.Inflammopharmacology. 2023 Oct;31(5):2303-2310. doi: 10.1007/s10787-023-01270-5. Epub 2023 Jul 5. Inflammopharmacology. 2023. PMID: 37405587 Review.
References
-
- Bosco M.C., Pierobon D., Blengio F., Raggi F., Vanni C., Gattorno M., et al. Hypoxia modulates the gene expression profile of immunoregulatory receptors in human mature dendritic cells: identification of TREM-1 as a novel hypoxic marker in vitro and in vivo. Blood. 2011;117:2625–2639. - PubMed
-
- Bouchon A., Dietrich J., Colonna M. Cutting edge: inflammatory responses can be triggered by TREM-1, a novel receptor expressed on neutrophils and monocytes. J Immunol. 1950;164(2000):4991–4995. - PubMed
-
- Gingras M.C., Lapillonne H., Margolin J.F. TREM-1, MDL-1, and DAP12 expression is associated with a mature stage of myeloid development. Mol Immunol. 2002;38:817–824. - PubMed
-
- de Oliveira Matos A., Dos Santos Dantas P.H., Figueira Marques Silva-Sales M., Sales-Campos H. The role of the triggering receptor expressed on myeloid cells-1 (TREM-1) in non-bacterial infections. Crit Rev Microbiol. 2020;46:237–252. - PubMed
-
- Matos A.O., Dantas P., Silva-Sales M., Sales-Campos H. TREM-1 isoforms in bacterial infections: to immune modulation and beyond. Crit Rev Microbiol. 2021;47:290–306. - PubMed
LinkOut - more resources
Full Text Sources
