Adaptive immune receptor genotyping using the corecount program
- PMID: 37114042
- PMCID: PMC10126697
- DOI: 10.3389/fimmu.2023.1125884
Adaptive immune receptor genotyping using the corecount program
Abstract
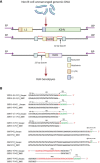
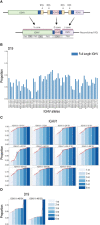
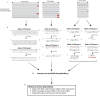
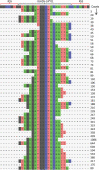
We present a new Rep-Seq analysis tool called corecount, for analyzing genotypic variation in immunoglobulin (IG) and T cell receptor (TCR) genes. corecount is highly efficient at identifying V alleles, including those that are infrequently used in expressed repertoires and those that contain 3' end variation that are otherwise refractory to reliable identification during germline inference from expressed libraries. Furthermore, corecount facilitates accurate D and J gene genotyping. The output is highly reproducible and facilitates the comparison of genotypes from multiple individuals, such as those from clinical cohorts. Here, we applied corecount to the genotypic analysis of IgM libraries from 16 individuals. To demonstrate the accuracy of corecount, we Sanger sequenced all the heavy chain IG alleles (65 IGHV, 27 IGHD and 7 IGHJ) from one individual from whom we also produced two independent IgM Rep-seq datasets. Genomic analysis revealed that 5 known IGHV and 2 IGHJ sequences are truncated in current reference databases. This dataset of genomically validated alleles and IgM libraries from the same individual provides a useful resource for benchmarking other bioinformatic programs that involve V, D and J assignments and germline inference, and may facilitate the development of AIRR-Seq analysis tools that can take benefit from the availability of more comprehensive reference databases.
Keywords: IGH; VDJ germline genes; genotyping; immune repertoires; inference.
Copyright © 2023 Narang, Kaduk, Chernyshev, Karlsson Hedestam and Corcoran.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
AIRR-C IG Reference Sets: curated sets of immunoglobulin heavy and light chain germline genes.Front Immunol. 2024 Feb 9;14:1330153. doi: 10.3389/fimmu.2023.1330153. eCollection 2023. Front Immunol. 2024. PMID: 38406579 Free PMC article.
-
Inferred Allelic Variants of Immunoglobulin Receptor Genes: A System for Their Evaluation, Documentation, and Naming.Front Immunol. 2019 Mar 18;10:435. doi: 10.3389/fimmu.2019.00435. eCollection 2019. Front Immunol. 2019. PMID: 30936866 Free PMC article. Review.
-
High-Quality Library Preparation for NGS-Based Immunoglobulin Germline Gene Inference and Repertoire Expression Analysis.Front Immunol. 2019 Apr 5;10:660. doi: 10.3389/fimmu.2019.00660. eCollection 2019. Front Immunol. 2019. PMID: 31024532 Free PMC article.
-
Automated analysis of high-throughput B-cell sequencing data reveals a high frequency of novel immunoglobulin V gene segment alleles.Proc Natl Acad Sci U S A. 2015 Feb 24;112(8):E862-70. doi: 10.1073/pnas.1417683112. Epub 2015 Feb 9. Proc Natl Acad Sci U S A. 2015. PMID: 25675496 Free PMC article.
-
Individual variation in the germline Ig gene repertoire inferred from variable region gene rearrangements.J Immunol. 2010 Jun 15;184(12):6986-92. doi: 10.4049/jimmunol.1000445. Epub 2010 May 21. J Immunol. 2010. PMID: 20495067 Free PMC article.
Cited by
-
Human immunoglobulin gene allelic variation impacts germline-targeting vaccine priming.NPJ Vaccines. 2024 Mar 11;9(1):58. doi: 10.1038/s41541-024-00811-5. NPJ Vaccines. 2024. PMID: 38467663 Free PMC article.
-
AIRR-C IG Reference Sets: curated sets of immunoglobulin heavy and light chain germline genes.Front Immunol. 2024 Feb 9;14:1330153. doi: 10.3389/fimmu.2023.1330153. eCollection 2023. Front Immunol. 2024. PMID: 38406579 Free PMC article.
References
-
- Watson CT, Steinberg KM, Huddleston J, Warren RL, Malig M, Schein J, et al. . Complete haplotype sequence of the human immunoglobulin heavy-chain variable, diversity, and joining genes and characterization of allelic and copy-number variation. Am J Hum Genet (2013) 92(4):530–46. doi: 10.1016/j.ajhg.2013.03.004 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

