Tuning the Biological Activity of PI3K δ Inhibitor by the Introduction of a Fluorine Atom Using the Computational Workflow
- PMID: 37110764
- PMCID: PMC10145010
- DOI: 10.3390/molecules28083531
Tuning the Biological Activity of PI3K δ Inhibitor by the Introduction of a Fluorine Atom Using the Computational Workflow
Abstract
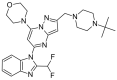
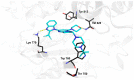
As a member of the class I PI3K family, phosphoinositide 3-kinase δ (PI3Kδ) is an important signaling biomolecule that controls immune cell differentiation, proliferation, migration, and survival. It also represents a potential and promising therapeutic approach for the management of numerous inflammatory and autoimmune diseases. We designed and assessed the biological activity of new fluorinated analogues of CPL302415, taking into account the therapeutic potential of our selective PI3K inhibitor and fluorine introduction as one of the most frequently used modifications of a lead compound to further improve its biological activity. In this paper, we compare and evaluate the accuracy of our previously described and validated in silico workflow with that of the standard (rigid) molecular docking approach. The findings demonstrated that a properly fitted catalytic (binding) pocket for our chemical cores at the induced-fit docking (IFD) and molecular dynamics (MD) stages, along with QM-derived atomic charges, can be used for activity prediction to better distinguish between active and inactive molecules. Moreover, the standard approach seems to be insufficient to score the halogenated derivatives due to the fixed atomic charges, which do not consider the response and indictive effects caused by fluorine. The proposed computational workflow provides a computational tool for the rational design of novel halogenated drugs.
Keywords: CPL302415; MD; MM-GBSA; PI3Kδ; QPLD; asthma; fluorine; induced-fit docking; molecular docking.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
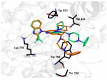

Similar articles
-
Theoretical studies on the selectivity mechanisms of PI3Kδ inhibition with marketed idelalisib and its derivatives by 3D-QSAR, molecular docking, and molecular dynamics simulation.J Mol Model. 2019 Jul 23;25(8):242. doi: 10.1007/s00894-019-4129-x. J Mol Model. 2019. PMID: 31338599
-
Identification of novel PI3Kδ inhibitors by docking, ADMET prediction and molecular dynamics simulations.Comput Biol Chem. 2019 Feb;78:190-204. doi: 10.1016/j.compbiolchem.2018.12.002. Epub 2018 Dec 7. Comput Biol Chem. 2019. PMID: 30557817
-
Isomeric Activity Cliffs-A Case Study for Fluorine Substitution of Aminergic G Protein-Coupled Receptor Ligands.Molecules. 2023 Jan 4;28(2):490. doi: 10.3390/molecules28020490. Molecules. 2023. PMID: 36677547 Free PMC article.
-
Small molecule inhibitors of phosphoinositide 3-kinase (PI3K) delta and gamma.Curr Top Med Chem. 2009;9(8):738-53. doi: 10.2174/156802609789044434. Curr Top Med Chem. 2009. PMID: 19689378 Review.
-
PI3K pathway defects leading to immunodeficiency and immune dysregulation.J Allergy Clin Immunol. 2019 May;143(5):1676-1687. doi: 10.1016/j.jaci.2019.03.017. J Allergy Clin Immunol. 2019. PMID: 31060715 Review.
References
-
- Haselmayer P., Camps M., Muzerelle M., El Bawab S., Waltzinger C., Bruns L., Abla N., Polokoff M.A., Jond-Necand C., Gaudet M., et al. Characterization of Novel PI3Kδ Inhibitors as Potential Therapeutics for SLE and Lupus Nephritis in Pre-Clinical Studies. Front. Immunol. 2014;5:233. doi: 10.3389/fimmu.2014.00233. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

